E14.5 cortex
library(COTAN)
library(data.table)
library(Matrix)
library(ggrepel)
#> Loading required package: ggplot2
# library(latex2exp)mycolours <- c(A = "#8491B4B2", B = "#E64B35FF")
my_theme = theme(axis.text.x = element_text(size = 14,
angle = 0, hjust = 0.5, vjust = 0.5,
face = "plain", colour = "#3C5488FF"),
axis.text.y = element_text(size = 14,
angle = 0, hjust = 0, vjust = 0.5,
face = "plain", colour = "#3C5488FF"),
axis.title.x = element_text(size = 14,
angle = 0, hjust = 0.5, vjust = 0,
face = "plain", colour = "#3C5488FF"),
axis.title.y = element_text(size = 14,
angle = 90, hjust = 0.5, vjust = 0.5,
face = "plain", colour = "#3C5488FF"))data = as.data.frame(fread(paste(data_dir,
"GSE123335_E14_combined_matrix.txt.gz",
sep = "/")))
data = as.data.frame(data)
rownames(data) = data$Gene
data = data[, 2:ncol(data)]
data[1:10, 1:10]
#> e14-WT10_AAAAAGCAAGAA e14-WT10_AAAAATCTCTCC e14-WT10_AAAACACATTCC
#> 0610005C13Rik 0 0 0
#> 0610007N19Rik 0 0 0
#> 0610007P14Rik 0 1 0
#> 0610009B14Rik 0 0 0
#> 0610009B22Rik 0 0 0
#> 0610009D07Rik 1 0 0
#> 0610009E02Rik 0 0 0
#> 0610009L18Rik 0 0 0
#> 0610009O20Rik 0 0 0
#> 0610010F05Rik 0 0 0
#> e14-WT10_AAAACCGTGGAT e14-WT10_AAAACGCGCCGA e14-WT10_AAAAGAGCCGAG
#> 0610005C13Rik 0 0 0
#> 0610007N19Rik 0 0 0
#> 0610007P14Rik 0 0 0
#> 0610009B14Rik 0 0 0
#> 0610009B22Rik 0 0 0
#> 0610009D07Rik 0 0 1
#> 0610009E02Rik 0 0 0
#> 0610009L18Rik 0 0 0
#> 0610009O20Rik 0 0 0
#> 0610010F05Rik 1 1 0
#> e14-WT10_AAAATGGACTCA e14-WT10_AAAATGTCACAA e14-WT10_AAAATTCCGCTT
#> 0610005C13Rik 0 0 0
#> 0610007N19Rik 0 0 0
#> 0610007P14Rik 0 0 0
#> 0610009B14Rik 0 0 0
#> 0610009B22Rik 0 0 1
#> 0610009D07Rik 2 0 1
#> 0610009E02Rik 0 0 0
#> 0610009L18Rik 0 0 0
#> 0610009O20Rik 0 0 0
#> 0610010F05Rik 0 3 0
#> e14-WT10_AAAATTGCAGAG
#> 0610005C13Rik 0
#> 0610007N19Rik 0
#> 0610007P14Rik 0
#> 0610009B14Rik 0
#> 0610009B22Rik 1
#> 0610009D07Rik 1
#> 0610009E02Rik 0
#> 0610009L18Rik 0
#> 0610009O20Rik 0
#> 0610010F05Rik 0Define a directory where the ouput will be stored.
0.1 Analytical pipeline
Initialize the COTAN object with the row count table and the metadata for the experiment.
obj = new("scCOTAN", raw = data)
obj = initRaw(obj, GEO = "GSE123335", sc.method = "10X",
cond = "mouse cortex E14.5")
#> [1] "Initializing S4 object"Now we can start the cleaning. Analysis requires and starts from a matrix of raw UMI counts after removing possible cell doublets or multiplets and low quality or dying cells (with too high mtRNA percentage, easily done with Seurat or other tools).
If we do not want to consider the mithocondrial genes we can remove them before starting the analysis.
genes_to_rem = rownames(obj@raw[grep("^mt",
rownames(obj@raw)), ]) #genes to remove : mithocondrial
obj@raw = obj@raw[!rownames(obj@raw) %in%
genes_to_rem, ]
cells_to_rem = colnames(obj@raw[which(colSums(obj@raw) ==
0)])
obj@raw = obj@raw[, !colnames(obj@raw) %in%
cells_to_rem]We want also to define a prefix to identify the sample.
0.2 Data cleaning
First, we create the directory to store all information regarding the data cleaning.
if (!file.exists(out_dir)) {
dir.create(file.path(out_dir))
}
if (!file.exists(paste(out_dir, "cleaning",
sep = ""))) {
dir.create(file.path(out_dir, "cleaning"))
}ttm = clean(obj)
#> [1] "Start estimation mu with linear method"
#> [1] 13719 11069
#> rowname e14-WT9-2_ACGCAACCACCA e14-WT9-2_GGTATGACCTCG
#> 5790 Hbb-y 1897.179627 1947.564248
#> 5784 Hba-a1 343.616508 341.769480
#> 5786 Hba-x 204.087380 311.766815
#> 5785 Hba-a2 214.500002 163.057958
#> 5788 Hbb-bs 220.747575 136.968685
#> 5789 Hbb-bt 112.456312 182.624913
#> 5787 Hbb-bh1 85.383496 7.826782
#> 4528 Fth1 37.485437 50.874083
#> 5604 Gpx1 22.907767 5.217855
#> 8291 Nudt4 8.330097 10.435709
#> 787 Actb 8.330097 1.304464
#> 10287 Rps9 2.082524 6.522318
#> 12647 Ubb 2.082524 3.913391
#> 1484 Atp5j2 4.165049 1.304464
#> 6044 Hsp90ab1 4.165049 1.304464
obj = ttm$object
ttm$pca.cell.2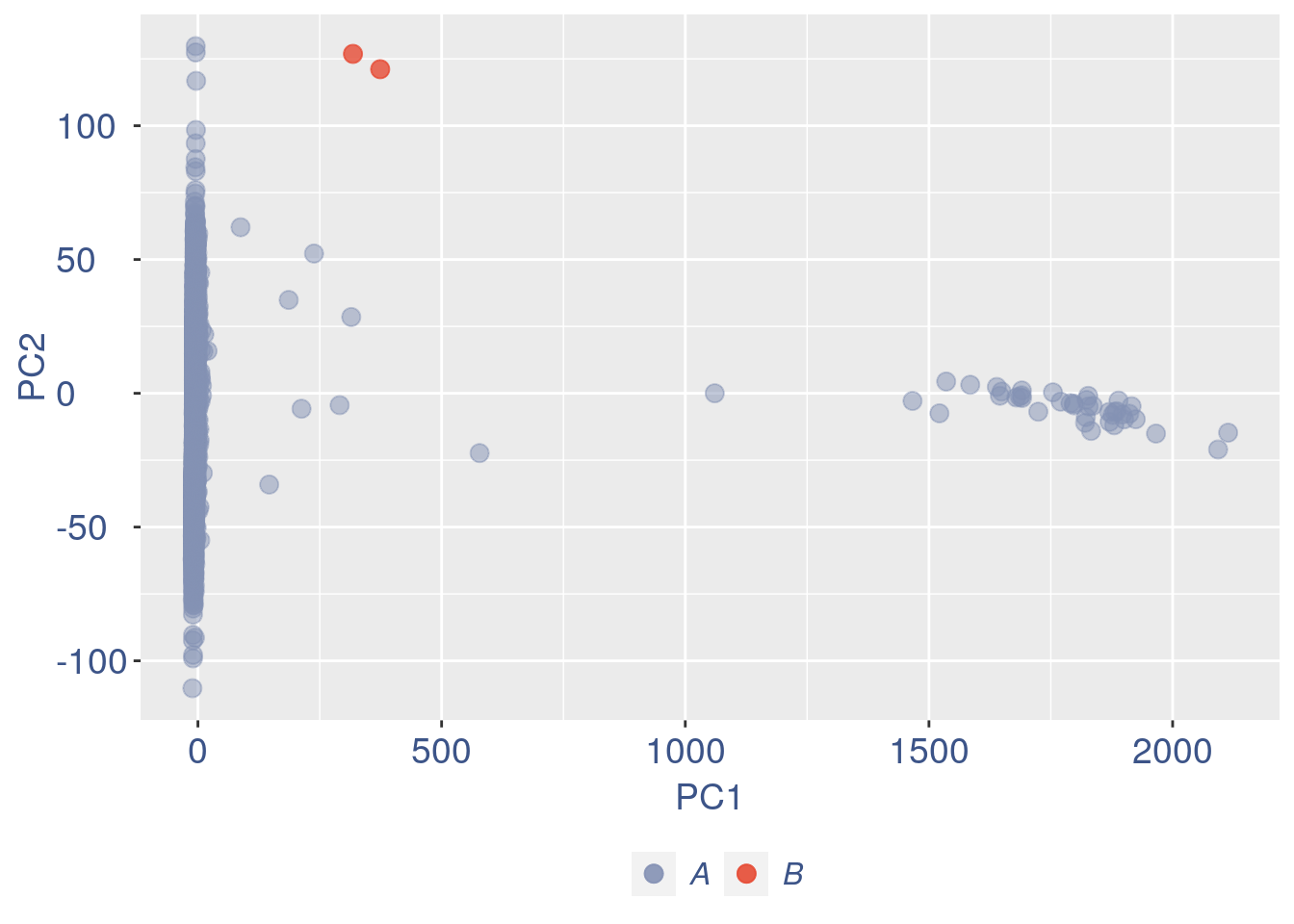
Run this when B cells need to be removed.
pdf(paste(out_dir, "cleaning/", t, "_", n_it,
"_plots_before_cells_exlusion.pdf", sep = ""))
ttm$pca.cell.2
ggplot(ttm$D, aes(x = n, y = means)) + geom_point() +
geom_text_repel(data = subset(ttm$D,
n > (max(ttm$D$n) - 15)), aes(n,
means, label = rownames(ttm$D[ttm$D$n >
(max(ttm$D$n) - 15), ])), nudge_y = 0.05,
nudge_x = 0.05, direction = "x",
angle = 90, vjust = 0, segment.size = 0.2) +
ggtitle("B cell group genes mean expression") +
my_theme + theme(plot.title = element_text(color = "#3C5488FF",
size = 20, face = "italic", vjust = -5,
hjust = 0.02))
dev.off()
#> png
#> 2
if (length(ttm$cl1) < length(ttm$cl2)) {
to_rem = ttm$cl1
} else {
to_rem = ttm$cl2
}
n_it = n_it + 1
obj@raw = obj@raw[, !colnames(obj@raw) %in%
to_rem]
# obj@raw = obj@raw[rownames(obj@raw)
# %in% rownames(cells),colnames(obj@raw)
# %in% colnames(cells)]
gc()
#> used (Mb) gc trigger (Mb) max used (Mb)
#> Ncells 3769210 201.3 6027342 321.9 6027342 321.9
#> Vcells 1051938372 8025.7 1764697884 13463.6 1470511620 11219.2
ttm = clean(obj)
#> [1] "Start estimation mu with linear method"
#> [1] 13718 11067
#> rowname e14-WT10_CCATCCGTTGAT e14-WT11_AGTGAAATCCTA
#> 5787 Hbb-bs 2012.644045 2014.140334
#> 5784 Hba-a1 472.811617 343.563383
#> 5788 Hbb-bt 444.059559 599.804406
#> 5785 Hba-a2 309.883289 257.672537
#> 12559 Tuba1a 9.584019 2.863028
#> 8457 Pabpc1 0.000000 5.726056
#> 7064 Map1b 0.000000 2.863028
#> 10253 Rps2 0.000000 1.431514
#> 7037 Malat1 0.000000 0.000000
#> 787 Actb 3.194673 1.431514
#> 12209 Tmsb4x 3.194673 0.000000
#> 9283 Prdx2 0.000000 0.000000
#> 10272 Rps5 0.000000 2.863028
#> 4528 Fth1 0.000000 0.000000
#> 8101 Nnat 6.389346 0.000000
#> e14-WT8-1_AAAGAACTTAAG e14-WT8-2_CCATCAATAGAA e14-WT8-2_CGCTTCCTCCTC
#> 5787 1778.831015 1402.254203 1772.939306
#> 5784 482.094689 536.773118 411.098786
#> 5788 415.191752 244.782529 371.080320
#> 5785 403.385352 437.111659 266.789772
#> 12559 3.935467 13.987573 7.276085
#> 8457 11.806401 1.748447 0.000000
#> 7064 5.903200 5.245340 3.638042
#> 10253 7.870934 3.496893 0.000000
#> 7037 3.935467 6.993787 10.914127
#> 787 3.935467 8.742233 6.063404
#> 12209 0.000000 10.490680 6.063404
#> 9283 3.935467 6.993787 2.425362
#> 10272 1.967733 8.742233 3.638042
#> 4528 0.000000 6.993787 1.212681
#> 8101 0.000000 3.496893 4.850723
#> e14-WT8-2_CTAACTTAATAC e14-WT8-2_TAGTCAGTTCAT
#> 5787 1966.495934 1531.577462
#> 5784 374.704813 512.110074
#> 5788 307.088907 430.124935
#> 5785 253.559648 367.150842
#> 12559 5.634659 3.564571
#> 8457 0.000000 15.446476
#> 7064 14.086647 0.000000
#> 10253 16.903977 1.188190
#> 7037 5.634659 2.376381
#> 787 2.817329 1.188190
#> 12209 5.634659 0.000000
#> 9283 5.634659 5.940952
#> 10272 5.634659 0.000000
#> 4528 0.000000 13.070095
#> 8101 2.817329 3.564571
# ttm = clean.sqrt(obj, cells)
obj = ttm$object
ttm$pca.cell.2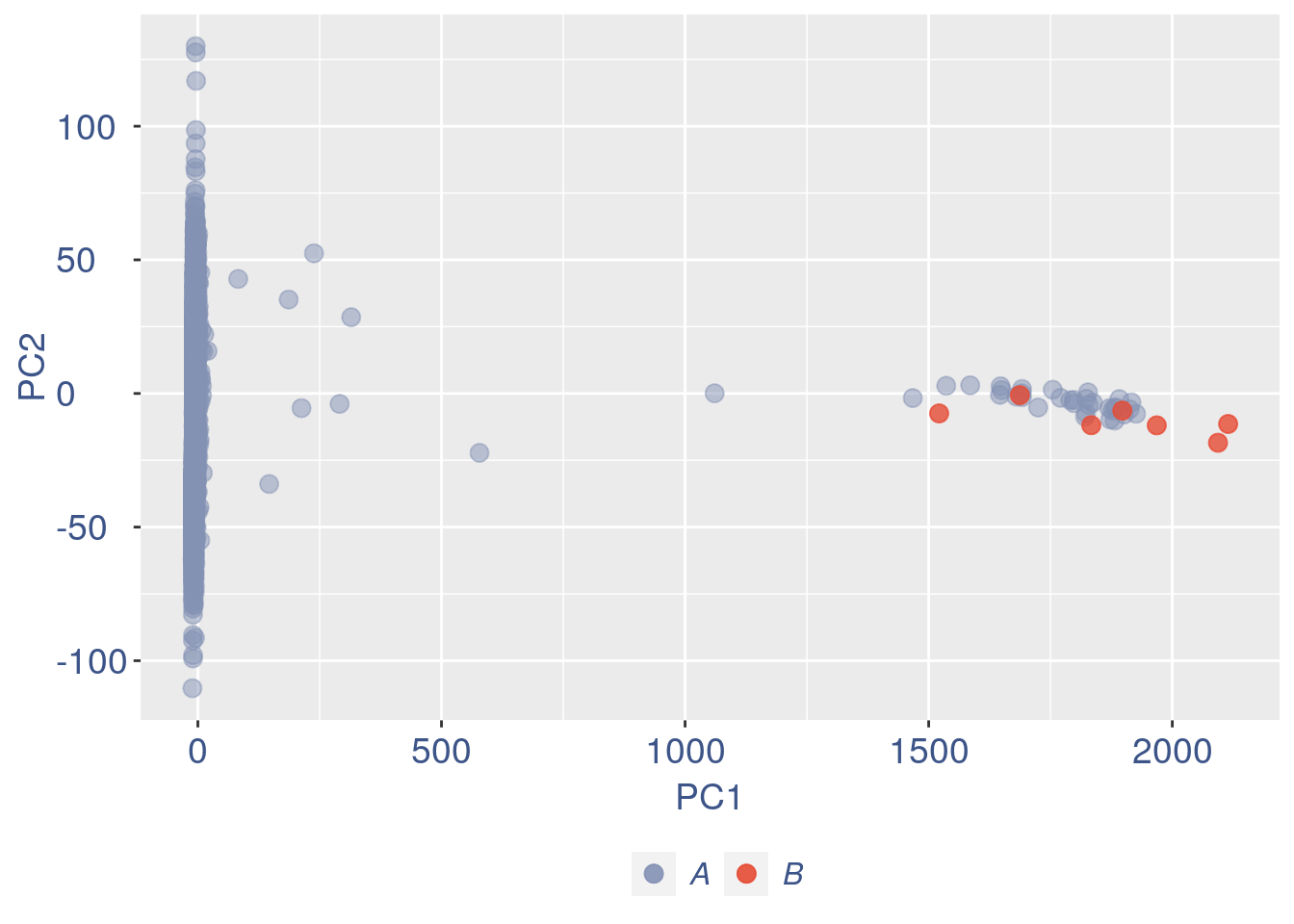
#> png
#> 2
#> used (Mb) gc trigger (Mb) max used (Mb)
#> Ncells 3769470 201.4 6027342 321.9 6027342 321.9
#> Vcells 1083135164 8263.7 2541340952 19388.9 2541334820 19388.9
#> [1] "Start estimation mu with linear method"
#> [1] 13718 11060
#> rowname e14-WT10_AGGGCATTTGTG e14-WT10_CGTATGCACTCG
#> 12554 Ttr 574.600871 1152.508633
#> 7037 Malat1 23.335319 24.379990
#> 6156 Igfbp2 16.435090 24.749384
#> 6043 Hsp90ab1 10.036696 5.540907
#> 787 Actb 6.649311 11.081814
#> 1855 Bsg 17.815136 20.686052
#> 11255 Sostdc1 8.907568 20.686052
#> 6433 Kcne2 10.413073 9.604239
#> 2817 Cox4i1 7.151146 8.126663
#> 10998 Slc6a15 13.047705 7.387876
#> 10244 Rps14 6.398394 8.496057
#> 4528 Fth1 7.652981 3.693938
#> 4055 Ezr 8.405733 2.955150
#> 2644 Clu 8.907568 8.865451
#> 10272 Rps5 3.638302 4.802119
#> e14-WT10_GTTATACAATGC e14-WT10_TAAAGATTGTAA e14-WT11_ACCTGTCTCAGA
#> 12554 1057.088055 370.823007 1203.605551
#> 7037 30.559707 41.440721 26.728791
#> 6156 12.501698 5.001466 27.538754
#> 6043 1.389078 22.863846 8.504615
#> 787 11.112621 5.715962 5.669744
#> 1855 12.501698 7.859447 22.678974
#> 11255 22.225242 7.859447 10.124542
#> 6433 13.890776 3.572476 10.934505
#> 2817 9.723543 5.001466 9.719560
#> 10998 12.501698 5.001466 12.554432
#> 10244 8.334466 7.144952 3.644835
#> 4528 2.778155 5.001466 4.454798
#> 4055 8.334466 2.143486 4.454798
#> 2644 11.112621 3.572476 11.744469
#> 10272 5.556310 6.430457 2.024908
#> e14-WT8-1_TGGCGGATGAGA e14-WT9-1_CGTGAATAGTAC e14-WT9-1_GACGTTTTTCAG
#> 12554 717.241538 622.721335 536.027646
#> 7037 14.533224 14.663768 11.245335
#> 6156 17.762830 5.865507 3.748445
#> 6043 11.303619 7.038608 18.742225
#> 787 6.997478 7.234125 11.245335
#> 1855 12.380154 5.865507 3.748445
#> 11255 9.957950 4.105855 0.000000
#> 6433 10.765351 13.490666 14.993780
#> 2817 9.688816 8.211710 22.490670
#> 10998 11.841887 13.686183 0.000000
#> 10244 9.688816 8.602744 7.496890
#> 4528 8.881415 7.625159 3.748445
#> 4055 7.804880 8.602744 7.496890
#> 2644 6.459211 2.346203 0.000000
#> 10272 4.306141 2.541720 26.239116
#> e14-WT9-1_GCCACCCTGAAC e14-WT9-1_GGTGCGAAAATC e14-WT9-2_CCGCGTACAAAG
#> 12554 417.3591554 387.243703 457.884548
#> 7037 20.4724669 67.556534 14.907869
#> 6156 13.0279335 12.063667 2.129696
#> 6043 14.4237835 9.650933 14.907869
#> 787 22.3336003 21.714600 10.648478
#> 1855 4.1875501 4.825467 2.129696
#> 11255 16.2849169 3.619100 0.000000
#> 6433 0.4652833 2.412733 10.648478
#> 2817 6.0486834 4.825467 6.389087
#> 10998 2.3264167 6.031833 12.778173
#> 10244 6.5139668 16.889133 8.518782
#> 4528 6.9792501 8.444567 17.037565
#> 4055 10.2362335 3.619100 8.518782
#> 2644 5.5834001 12.063667 0.000000
#> 10272 6.5139668 6.031833 2.129696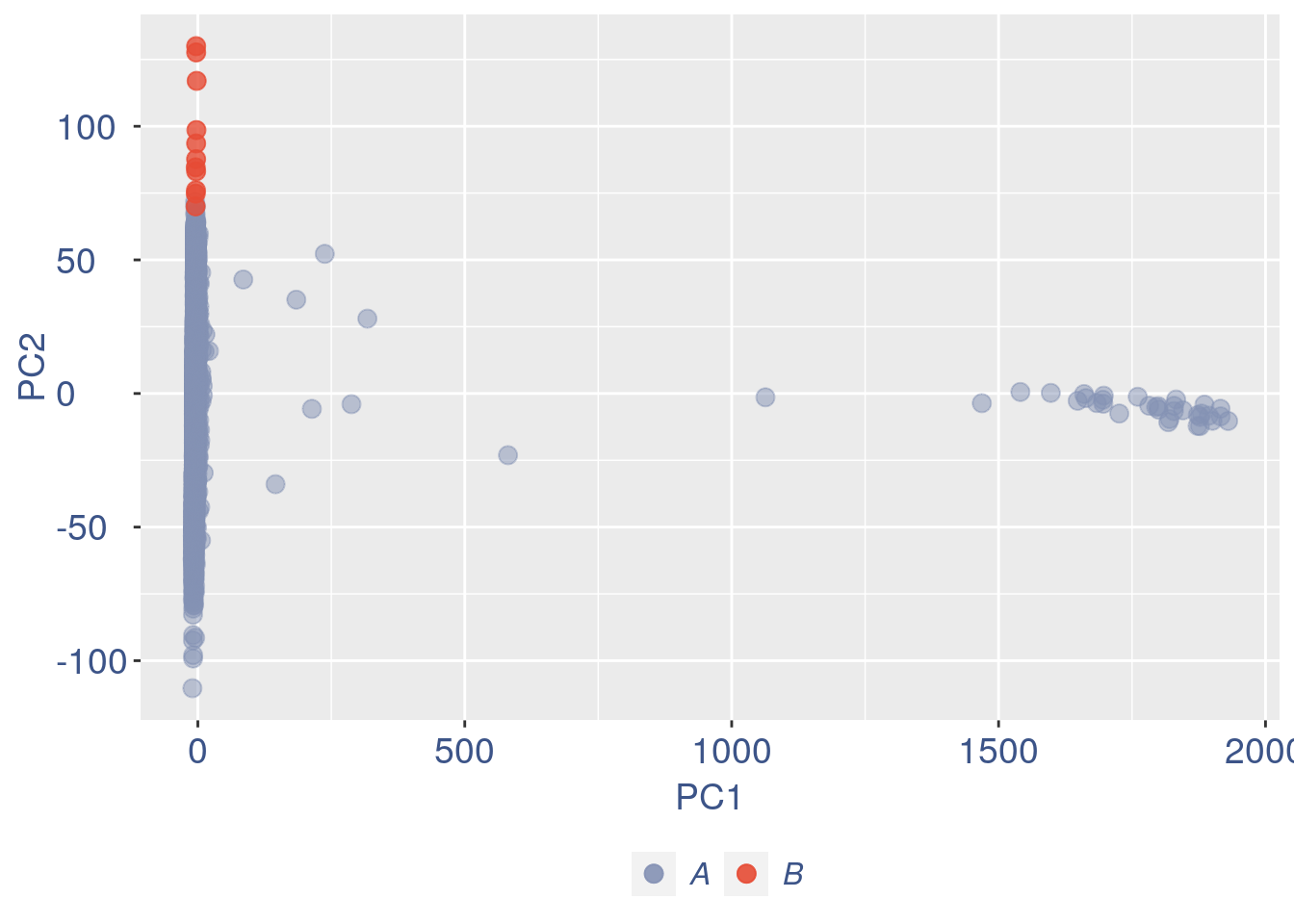
#> png
#> 2
#> used (Mb) gc trigger (Mb) max used (Mb)
#> Ncells 3769701 201.4 6027342 321.9 6027342 321.9
#> Vcells 1082653537 8260.0 3049689142 23267.3 3049682474 23267.3
#> [1] "Start estimation mu with linear method"
#> [1] 13690 11049
#> rowname e14-WT10_ATCGTAAGTTGA e14-WT10_CGTGCCTGAGCC e14-WT10_CGTGGAAAGTAC
#> 5775 Hbb-bs 1682.041404 1290.677184 1323.494152
#> 5776 Hbb-bt 847.748868 587.152326 925.728288
#> 5772 Hba-a1 341.454405 314.734805 387.514167
#> 5773 Hba-a2 290.993163 237.153116 277.821003
#> 8441 Pabpc1 0.000000 6.171271 14.352377
#> 5592 Gpx1 5.046124 15.868982 62.535355
#> 4517 Fth1 8.410207 22.921863 16.402716
#> 993 Alas2 5.046124 6.171271 14.352377
#> 12534 Tuba1a 3.364083 8.816101 4.100679
#> 9265 Prdx2 1.682041 7.052881 9.226528
#> 11116 Snca 0.000000 7.052881 4.100679
#> 786 Actb 1.682041 1.763220 3.075509
#> 7402 Mkrn1 0.000000 2.644830 3.075509
#> 12184 Tmsb4x 5.046124 4.408050 3.075509
#> 7022 Malat1 1.682041 11.460931 2.050340
#> e14-WT10_CTAAAGCGTCTG e14-WT10_CTTTTTCCCCCA e14-WT11_CAATTAGCTGTC
#> 5775 1592.232384 1629.3932719 1402.5033117
#> 5776 904.520565 713.0455036 639.0611420
#> 5772 411.522334 466.1076310 444.6264322
#> 5773 294.141802 372.8861048 402.4512561
#> 8441 2.761895 0.9917184 38.6010086
#> 5592 11.047579 0.0000000 28.5933397
#> 4517 9.666632 0.0000000 4.2890010
#> 993 4.142842 3.9668735 15.0115033
#> 12534 4.142842 10.9089020 4.2890010
#> 9265 4.142842 0.0000000 2.8593340
#> 11116 0.000000 0.0000000 5.7186679
#> 786 1.380947 1.9834367 4.2890010
#> 7402 0.000000 0.0000000 2.8593340
#> 12184 1.380947 0.9917184 3.5741675
#> 7022 0.000000 0.9917184 0.7148335
#> e14-WT11_CTAGGCGCTTCA e14-WT11_CTCCTAACGGTN e14-WT11_GATTGCTGCGCT
#> 5775 1549.5086779 1249.031355 1660.377269
#> 5776 706.5077105 850.505421 720.472075
#> 5772 462.9913317 437.131278 409.608489
#> 5773 366.0500981 374.768799 305.987294
#> 8441 13.1840078 30.290347 3.657219
#> 5592 3.8776493 35.041774 4.876292
#> 4517 9.3063584 21.381421 9.752583
#> 993 6.2042390 20.193564 9.752583
#> 12534 3.1021195 2.969642 6.095364
#> 9265 3.1021195 5.939284 3.657219
#> 11116 6.9797688 7.721069 7.314437
#> 786 1.5510597 4.157499 1.219073
#> 7402 6.2042390 2.375713 7.314437
#> 12184 3.1021195 2.375713 0.000000
#> 7022 0.7755299 4.157499 0.000000
#> e14-WT11_GCATACAAGATC e14-WT11_GCGTCGTCGCTC e14-WT11_GGTCGCTTCCTT
#> 5775 1692.610503 1436.258409 1544.013186
#> 5776 723.937556 734.401014 883.994055
#> 5772 362.824496 419.812692 391.398041
#> 5773 335.441515 317.842684 327.405206
#> 8441 1.711436 28.204470 17.858466
#> 5592 0.000000 24.950108 3.720514
#> 4517 1.711436 10.847873 5.952822
#> 993 1.711436 18.441384 8.185130
#> 12534 6.845745 1.084787 2.976411
#> 9265 0.000000 5.423937 1.488205
#> 11116 10.268618 7.593511 5.208719
#> 786 1.711436 2.169575 1.488205
#> 7402 0.000000 5.423937 4.464616
#> 12184 0.000000 4.339149 1.488205
#> 7022 0.000000 1.084787 0.000000
#> e14-WT11_GTTCAAAGGCGC e14-WT11_TGTTACTTTTGT e14-WT11_TTATACACTGAT
#> 5775 1629.473121 1416.9124496 1627.893047
#> 5776 771.608061 715.3898781 633.436856
#> 5772 417.170212 447.8324323 412.593526
#> 5773 302.683650 353.2084575 341.183108
#> 8441 31.366181 19.5773741 30.415548
#> 5592 0.000000 40.7861960 14.546567
#> 4517 3.136618 13.8673066 1.322415
#> 993 6.273236 16.3144784 9.256906
#> 12534 4.704927 5.7100674 7.934491
#> 9265 1.568309 2.4471718 1.322415
#> 11116 6.273236 4.0786196 3.967245
#> 786 3.136618 0.8157239 0.000000
#> 7402 3.136618 5.7100674 10.579321
#> 12184 0.000000 1.6314478 2.644830
#> 7022 3.136618 4.0786196 2.644830
#> e14-WT11_TTCATAAGCTCT e14-WT11_TTGCTTATTCCN e14-WT8-1_AAGTTTCAGCCG
#> 5775 1664.809667 1418.0806178 1519.291499
#> 5776 649.978450 729.9503448 696.721912
#> 5772 309.449199 462.5553400 373.895267
#> 5773 256.748244 363.0742601 244.399833
#> 8441 13.513065 32.3155101 14.591035
#> 5592 0.000000 9.5045618 14.591035
#> 4517 10.810452 17.1082112 1.823879
#> 993 0.000000 6.9700120 1.823879
#> 12534 6.756533 5.0690996 7.295517
#> 9265 2.702613 8.2372869 1.823879
#> 11116 1.351307 5.7027371 1.823879
#> 786 5.405226 1.9009124 1.823879
#> 7402 1.351307 3.1681873 3.647759
#> 12184 1.351307 1.2672749 3.647759
#> 7022 2.702613 0.6336375 1.823879
#> e14-WT8-1_CACTAAGTCTCC e14-WT8-1_CTACACACTGAT e14-WT8-1_GAAACCCGGTCG
#> 5775 1689.564912 1420.433130 1708.064242
#> 5776 719.271920 745.464050 620.898004
#> 5772 431.241330 426.210968 409.174164
#> 5773 352.394967 367.870303 309.259542
#> 8441 1.609109 21.877749 14.273517
#> 5592 12.872876 39.704064 14.273517
#> 4517 17.700204 22.688036 26.168115
#> 993 9.654657 14.585166 9.515678
#> 12534 0.000000 1.620574 4.757839
#> 9265 1.609109 11.344018 11.894598
#> 11116 9.654657 7.292583 0.000000
#> 786 1.609109 4.861722 0.000000
#> 7402 3.218219 5.672009 2.378920
#> 12184 0.000000 0.000000 7.136759
#> 7022 6.436438 0.810287 2.378920
#> e14-WT8-1_TTACAATGACCA e14-WT8-1_TTCGCATAGGTT e14-WT8-2_AACGATTAGTCN
#> 5775 889.783883 486.633003 1220.913353
#> 5776 433.349934 264.728354 628.747513
#> 5772 322.651585 150.531809 617.315740
#> 5773 249.202444 121.723135 472.132224
#> 8441 12.591281 3.114451 3.429532
#> 5592 9.443461 19.984395 1.143177
#> 4517 11.542008 7.267053 2.286355
#> 993 4.197094 4.412139 11.431773
#> 12534 15.739102 48.793069 4.572709
#> 9265 8.394188 9.602891 1.143177
#> 11116 3.672457 2.076301 4.572709
#> 786 14.689828 20.243933 2.286355
#> 7402 2.098547 2.076301 3.429532
#> 12184 13.640555 14.015031 1.143177
#> 7022 6.295641 10.900579 4.572709
#> e14-WT8-2_CCTCTCGTCGGG e14-WT8-2_CTGGGGTGATGT e14-WT8-2_GGCACCGTATGG
#> 5775 1531.321089 1684.388974 1582.220151
#> 5776 793.822772 578.883084 670.116770
#> 5772 499.879528 554.762956 532.370545
#> 5773 362.588672 373.861992 362.979917
#> 8441 0.000000 34.170182 0.000000
#> 5592 15.841253 0.000000 1.861435
#> 4517 1.760139 2.010011 0.000000
#> 993 14.081113 6.030032 5.584306
#> 12534 1.760139 0.000000 1.861435
#> 9265 0.000000 4.020021 0.000000
#> 11116 3.520278 8.040043 0.000000
#> 786 1.760139 2.010011 0.000000
#> 7402 3.520278 2.010011 3.722871
#> 12184 0.000000 2.010011 1.861435
#> 7022 0.000000 0.000000 3.722871
#> e14-WT9-1_ATTATCACTACA e14-WT9-1_CTAGCTTTGAAA e14-WT9-1_GCAAACGTCTCG
#> 5775 1322.492721 1487.0921144 1529.286874
#> 5776 893.634999 793.3998616 773.555365
#> 5772 391.849695 384.3430049 398.066125
#> 5773 333.072241 345.1417228 365.983183
#> 8441 6.530828 36.6446767 15.447342
#> 5592 25.034842 35.7924750 4.753028
#> 4517 7.619300 15.3396321 8.317800
#> 993 6.530828 9.3742196 11.882571
#> 12534 1.088471 0.8522018 2.376514
#> 9265 5.442357 17.0440357 2.376514
#> 11116 1.088471 5.1132107 2.376514
#> 786 1.088471 2.5566054 8.317800
#> 7402 0.000000 5.9654125 1.188257
#> 12184 3.265414 0.0000000 3.564771
#> 7022 3.265414 0.0000000 3.564771
#> e14-WT9-2_AAACGTGGCAAT e14-WT9-2_CAATTATGAATC e14-WT9-2_GTCAACCAACAG
#> 5775 1678.506548 1471.861014 1560.455108
#> 5776 700.246520 939.174853 818.599401
#> 5772 403.119585 322.799843 393.020735
#> 5773 293.651767 285.604800 326.742090
#> 8441 0.000000 0.000000 41.860197
#> 5592 1.737584 3.985183 1.162783
#> 4517 13.900675 21.254311 11.627832
#> 993 10.425507 1.328394 8.139483
#> 12534 1.737584 1.328394 0.000000
#> 9265 5.212753 6.641972 4.651133
#> 11116 3.475169 0.000000 4.651133
#> 786 5.212753 6.641972 3.488350
#> 7402 1.737584 0.000000 4.651133
#> 12184 3.475169 0.000000 1.162783
#> 7022 1.737584 2.656789 0.000000
#> e14-WT9-2_GTCTAACAGTCN e14-WT9-2_TTTTAAGATGAG
#> 5775 1577.6789061 1666.845819
#> 5776 714.4532766 818.433289
#> 5772 401.3393709 368.744669
#> 5773 291.4900172 316.280996
#> 8441 8.6495554 23.983393
#> 5592 10.3794665 0.000000
#> 4517 23.3537996 4.496886
#> 993 10.3794665 7.494810
#> 12534 1.7299111 2.997924
#> 9265 3.4598222 2.997924
#> 11116 6.0546888 2.997924
#> 786 3.4598222 5.995848
#> 7402 6.0546888 10.492734
#> 12184 1.7299111 2.997924
#> 7022 0.8649555 0.000000
#> png
#> 2
#> used (Mb) gc trigger (Mb) max used (Mb)
#> Ncells 3769925 201.4 6027342 321.9 6027342 321.9
#> Vcells 1080752808 8245.5 3049689142 23267.3 3049682474 23267.3
#> [1] "Start estimation mu with linear method"
#> [1] 13686 11014
#> rowname e14-WT10_AGAGCTGGACGT e14-WT10_ATGTGTAGTCAC
#> 7250 Meg3 84.361212 77.714463
#> 7019 Malat1 62.160893 70.649512
#> 12530 Tuba1a 39.960574 14.129902
#> 7046 Map1b 0.000000 21.194854
#> 8082 Nnat 4.440064 7.064951
#> 11119 Snhg11 39.960574 0.000000
#> 12492 Ttc3 22.200319 14.129902
#> 785 Actb 13.320191 14.129902
#> 5609 Gria2 0.000000 28.259805
#> 5960 Hnrnpa2b1 17.760255 21.194854
#> 12180 Tmsb4x 0.000000 21.194854
#> 10321 Rtn1 22.200319 14.129902
#> 972 Akap9 0.000000 14.129902
#> 7050 Map2 8.880128 21.194854
#> 6027 Hsp90ab1 8.880128 7.064951
#> e14-WT10_CAGCGTCTGTTC e14-WT10_CATGGTAAGACC e14-WT10_CTGAATGCAGCG
#> 7250 68.956688 119.947404 66.998755
#> 7019 62.252565 119.947404 22.332918
#> 12530 31.605149 28.683075 33.499377
#> 7046 2.873195 5.215105 9.571251
#> 8082 16.281440 18.252866 19.142501
#> 11119 20.112367 65.188807 17.547293
#> 12492 10.535050 0.000000 17.547293
#> 785 14.365977 2.607552 13.559272
#> 5609 9.577318 13.037761 0.000000
#> 5960 10.535050 7.822657 1.595208
#> 12180 7.661854 10.430209 12.761668
#> 10321 12.450513 7.822657 19.142501
#> 972 2.873195 18.252866 3.190417
#> 7050 1.915464 2.607552 2.392813
#> 6027 8.619586 2.607552 10.368855
#> e14-WT10_GGCACATCAGCG e14-WT10_TACATACAATGG e14-WT10_TACGACGAATTA
#> 7250 139.998257 120.466294 131.6680
#> 7019 101.816914 102.750662 184.3352
#> 12530 0.000000 24.801884 26.3336
#> 7046 6.363557 7.086253 26.3336
#> 8082 12.727114 7.086253 16.4585
#> 11119 25.454229 0.000000 13.1668
#> 12492 0.000000 3.543126 6.5834
#> 785 6.363557 3.543126 3.2917
#> 5609 0.000000 7.086253 13.1668
#> 5960 6.363557 7.086253 3.2917
#> 12180 6.363557 3.543126 0.0000
#> 10321 6.363557 17.715631 6.5834
#> 972 0.000000 7.086253 9.8751
#> 7050 0.000000 3.543126 3.2917
#> 6027 0.000000 0.000000 6.5834
#> e14-WT10_TAGACCGCTTGA e14-WT10_TCGACTCATGTG e14-WT10_TTACGCAACCAT
#> 7250 35.565303 89.584426 53.415313
#> 7019 97.804583 39.522541 67.150679
#> 12530 120.032898 97.488935 83.938349
#> 7046 4.445663 7.904508 15.261518
#> 8082 4.445663 10.539344 9.156911
#> 11119 4.445663 0.000000 0.000000
#> 12492 8.891326 5.269672 3.052304
#> 785 4.445663 5.269672 10.683063
#> 5609 4.445663 5.269672 6.104607
#> 5960 22.228314 7.904508 7.630759
#> 12180 13.336989 10.539344 10.683063
#> 10321 4.445663 13.174180 7.630759
#> 972 4.445663 7.904508 6.104607
#> 7050 0.000000 2.634836 7.630759
#> 6027 0.000000 10.539344 4.578455
#> e14-WT10_TTTCCCCAATAA e14-WT11_ACGATGGTGCAG e14-WT11_TAACCACGGGTA
#> 7250 71.285877 91.496357 64.512359
#> 7019 27.218244 16.805453 22.769068
#> 12530 9.072748 54.150905 22.769068
#> 7046 9.072748 16.805453 53.127825
#> 8082 5.184427 22.407271 3.794845
#> 11119 0.000000 16.805453 0.000000
#> 12492 2.592214 1.867273 7.589689
#> 785 14.257175 22.407271 0.000000
#> 5609 1.296107 5.601818 11.384534
#> 5960 5.184427 5.601818 7.589689
#> 12180 15.553282 16.805453 3.794845
#> 10321 1.296107 13.070908 15.179379
#> 972 2.592214 0.000000 26.563912
#> 7050 5.184427 7.469090 18.974223
#> 6027 6.480534 9.336363 7.589689
#> e14-WT11_TACAAGAAGTCC e14-WT11_TGAAAAGCAGAG e14-WT8-1_AAAATCCATGGC
#> 7250 55.518278 119.025119 76.048920
#> 7019 17.664907 45.342902 59.752723
#> 12530 27.759139 17.003588 16.296197
#> 7046 8.832453 17.003588 10.864131
#> 8082 22.712023 22.671451 38.024460
#> 11119 35.329813 0.000000 21.728263
#> 12492 10.094232 5.667863 10.864131
#> 785 22.712023 17.003588 5.432066
#> 5609 2.523558 22.671451 5.432066
#> 5960 18.926686 0.000000 0.000000
#> 12180 11.356011 0.000000 5.432066
#> 10321 11.356011 0.000000 0.000000
#> 972 5.047116 5.667863 21.728263
#> 7050 1.261779 11.335726 16.296197
#> 6027 7.570674 0.000000 5.432066
#> e14-WT8-1_ACCCACACTATC e14-WT8-1_ATATCCATTACC e14-WT8-1_ATCCACCAGTGG
#> 7250 139.858973 50.005825 59.312903
#> 7019 110.922634 53.131189 44.484677
#> 12530 4.822723 65.632645 51.898790
#> 7046 4.822723 14.064138 14.828226
#> 8082 0.000000 3.125364 3.707056
#> 11119 28.936339 0.000000 0.000000
#> 12492 14.468170 10.938774 3.707056
#> 785 4.822723 7.813410 22.242339
#> 5609 28.936339 7.813410 0.000000
#> 5960 4.822723 3.125364 0.000000
#> 12180 9.645446 4.688046 7.414113
#> 10321 4.822723 7.813410 7.414113
#> 972 0.000000 3.125364 11.121169
#> 7050 4.822723 3.125364 0.000000
#> 6027 4.822723 6.250728 0.000000
#> e14-WT8-1_CCATAGTATTCG e14-WT8-1_GACACGGGACCC e14-WT8-1_GGCTACTGTGGT
#> 7250 128.085854 91.00924 103.337855
#> 7019 12.198653 52.68956 79.490657
#> 12530 6.099326 23.94980 7.949066
#> 7046 24.397306 4.78996 15.898131
#> 8082 6.099326 14.36988 3.974533
#> 11119 12.198653 0.00000 3.974533
#> 12492 6.099326 9.57992 15.898131
#> 785 0.000000 9.57992 7.949066
#> 5609 6.099326 14.36988 3.974533
#> 5960 0.000000 14.36988 11.923599
#> 12180 0.000000 23.94980 3.974533
#> 10321 0.000000 0.00000 0.000000
#> 972 12.198653 4.78996 11.923599
#> 7050 12.198653 0.00000 11.923599
#> 6027 0.000000 4.78996 27.821730
#> e14-WT8-1_GGGGACGTGCAA e14-WT8-1_TACATGATCGAT e14-WT8-1_TCGTTTAGCGGC
#> 7250 41.655972 211.276079 34.514510
#> 7019 21.924196 28.998678 49.306443
#> 12530 6.577259 16.570673 83.820953
#> 7046 0.000000 82.853364 9.861289
#> 8082 0.000000 0.000000 9.861289
#> 11119 0.000000 57.997355 0.000000
#> 12492 0.000000 16.570673 9.861289
#> 785 28.501454 0.000000 19.722577
#> 5609 4.384839 8.285336 0.000000
#> 5960 2.192420 4.142668 4.930644
#> 12180 4.384839 4.142668 14.791933
#> 10321 2.192420 8.285336 0.000000
#> 972 2.192420 0.000000 4.930644
#> 7050 0.000000 16.570673 9.861289
#> 6027 2.192420 12.428005 0.000000
#> e14-WT8-1_TGTCACTAAAAG e14-WT8-1_TTGGAGGCTATA e14-WT8-2_AAACCGATTGGG
#> 7250 81.621262 65.632645 64.512359
#> 7019 31.392793 32.816323 75.896893
#> 12530 25.114234 56.256553 0.000000
#> 7046 9.417838 18.752184 7.589689
#> 8082 25.114234 0.000000 7.589689
#> 11119 0.000000 0.000000 0.000000
#> 12492 3.139279 4.688046 7.589689
#> 785 9.417838 4.688046 3.794845
#> 5609 21.974955 0.000000 18.974223
#> 5960 12.557117 14.064138 15.179379
#> 12180 3.139279 0.000000 0.000000
#> 10321 12.557117 4.688046 0.000000
#> 972 3.139279 14.064138 7.589689
#> 7050 0.000000 9.376092 0.000000
#> 6027 3.139279 9.376092 0.000000
#> e14-WT8-2_AGTCTTCTTCAC e14-WT8-2_CCGACCTCATTG e14-WT8-2_CTGCTTGTACGA
#> 7250 110.983053 109.073479 51.440817
#> 7019 58.888967 167.505700 10.288163
#> 12530 38.504325 23.372888 61.728980
#> 7046 10.192321 7.790963 10.288163
#> 8082 5.662401 62.327702 17.146939
#> 11119 0.000000 11.686444 0.000000
#> 12492 20.384642 11.686444 13.717551
#> 785 3.397440 3.895481 27.435102
#> 5609 6.794881 11.686444 6.858776
#> 5960 4.529921 7.790963 10.288163
#> 12180 3.397440 0.000000 37.723266
#> 10321 6.794881 3.895481 20.576327
#> 972 10.192321 3.895481 0.000000
#> 7050 3.397440 7.790963 3.429388
#> 6027 11.324801 7.790963 10.288163
#> e14-WT8-2_GACCCTTTTCTA e14-WT8-2_TAACCACCCCGT e14-WT8-2_TACGTGGCGTCC
#> 7250 140.242674 111.595733 127.732270
#> 7019 99.137062 101.450666 106.443559
#> 12530 12.089886 15.217600 8.515485
#> 7046 29.015726 10.145067 51.092908
#> 8082 9.671909 10.145067 29.804196
#> 11119 19.343817 20.290133 4.257742
#> 12492 43.523588 0.000000 21.288712
#> 785 2.417977 5.072533 0.000000
#> 5609 21.761794 15.217600 8.515485
#> 5960 7.253931 25.362667 8.515485
#> 12180 0.000000 5.072533 0.000000
#> 10321 0.000000 15.217600 8.515485
#> 972 0.000000 5.072533 17.030969
#> 7050 16.925840 0.000000 21.288712
#> 6027 2.417977 0.000000 4.257742
#> e14-WT8-2_TCCTAGCCTGGT e14-WT9-1_CCGAATCGGACC e14-WT9-1_TAGGTCGGTGCG
#> 7250 203.652363 41.475420 75.008737
#> 7019 88.990949 41.475420 13.236836
#> 12530 5.134093 82.950839 39.710508
#> 7046 8.556822 13.825140 26.473672
#> 8082 5.134093 2.765028 4.412279
#> 11119 46.206839 0.000000 0.000000
#> 12492 25.670466 5.530056 13.236836
#> 785 0.000000 16.590168 0.000000
#> 5609 3.422729 11.060112 4.412279
#> 5960 3.422729 8.295084 0.000000
#> 12180 1.711364 11.060112 13.236836
#> 10321 6.845458 8.295084 8.824557
#> 972 15.402280 5.530056 17.649115
#> 7050 17.113644 11.060112 13.236836
#> 6027 6.845458 22.120224 0.000000
#> e14-WT9-2_GTATCTTACCAC e14-WT9-2_TGGTTCTGCACT e14-WT9-2_TTGTGATAGAGC
#> 7250 74.937874 94.406559 53.0668940
#> 7019 51.623869 40.459954 11.4978270
#> 12530 24.979291 26.973303 31.8401364
#> 7046 16.652861 62.038596 14.1511717
#> 8082 23.314005 10.789321 19.4578611
#> 11119 4.995858 21.578642 22.1112058
#> 12492 4.995858 16.183982 7.0755859
#> 785 29.975150 0.000000 26.5334470
#> 5609 1.665286 8.091991 0.8844482
#> 5960 3.330572 10.789321 3.5377929
#> 12180 9.991717 5.394661 8.8444823
#> 10321 3.330572 2.697330 7.9600341
#> 972 3.330572 5.394661 6.1911376
#> 7050 6.661144 5.394661 5.3066894
#> 6027 11.657003 21.578642 13.2667235
Run this only in the last iteration, instead the previous code, when B cells group has not to be removed.
pdf(paste(out_dir, "cleaning/", t, "_", n_it,
"_plots_before_cells_exlusion.pdf", sep = ""))
ttm$pca.cell.2
ggplot(ttm$D, aes(x = n, y = means)) + geom_point() +
geom_text_repel(data = subset(ttm$D,
n > (max(ttm$D$n) - 15)), aes(n,
means, label = rownames(ttm$D[ttm$D$n >
(max(ttm$D$n) - 15), ])), nudge_y = 0.05,
nudge_x = 0.05, direction = "x",
angle = 90, vjust = 0, segment.size = 0.2) +
ggtitle(label = "B cell group genes mean expression",
subtitle = " - B group NOT removed -") +
my_theme + theme(plot.title = element_text(color = "#3C5488FF",
size = 20, face = "italic", vjust = -10,
hjust = 0.02), plot.subtitle = element_text(color = "darkred",
vjust = -15, hjust = 0.01))
dev.off()
#> png
#> 2To colour the pca based on nu_j (so the cells’ efficiency).
nu_est = round(obj@nu, digits = 7)
plot.nu <- ggplot(ttm$pca_cells, aes(x = PC1,
y = PC2, colour = log(nu_est)))
plot.nu = plot.nu + geom_point(size = 1,
alpha = 0.8) + scale_color_gradient2(low = "#E64B35B2",
mid = "#4DBBD5B2", high = "#3C5488B2",
midpoint = log(mean(nu_est)), name = "ln (nu)") +
ggtitle("Cells PCA coloured by cells efficiency") +
my_theme + theme(plot.title = element_text(color = "#3C5488FF",
size = 20), legend.title = element_text(color = "#3C5488FF",
size = 14, face = "italic"), legend.text = element_text(color = "#3C5488FF",
size = 11), legend.key.width = unit(2,
"mm"), legend.position = "right")
pdf(paste(out_dir, "cleaning/", t, "_plots_PCA_efficiency_colored.pdf",
sep = ""))
plot.nu
dev.off()
#> png
#> 2
plot.nu
The next part is used to remove the cells with efficiency too low.
nu_df = data.frame(nu = sort(obj@nu), n = c(1:length(obj@nu)))
ggplot(nu_df, aes(x = n, y = nu)) + geom_point(colour = "#8491B4B2",
size = 1) + my_theme #+ ylim(0,1) + xlim(0,70)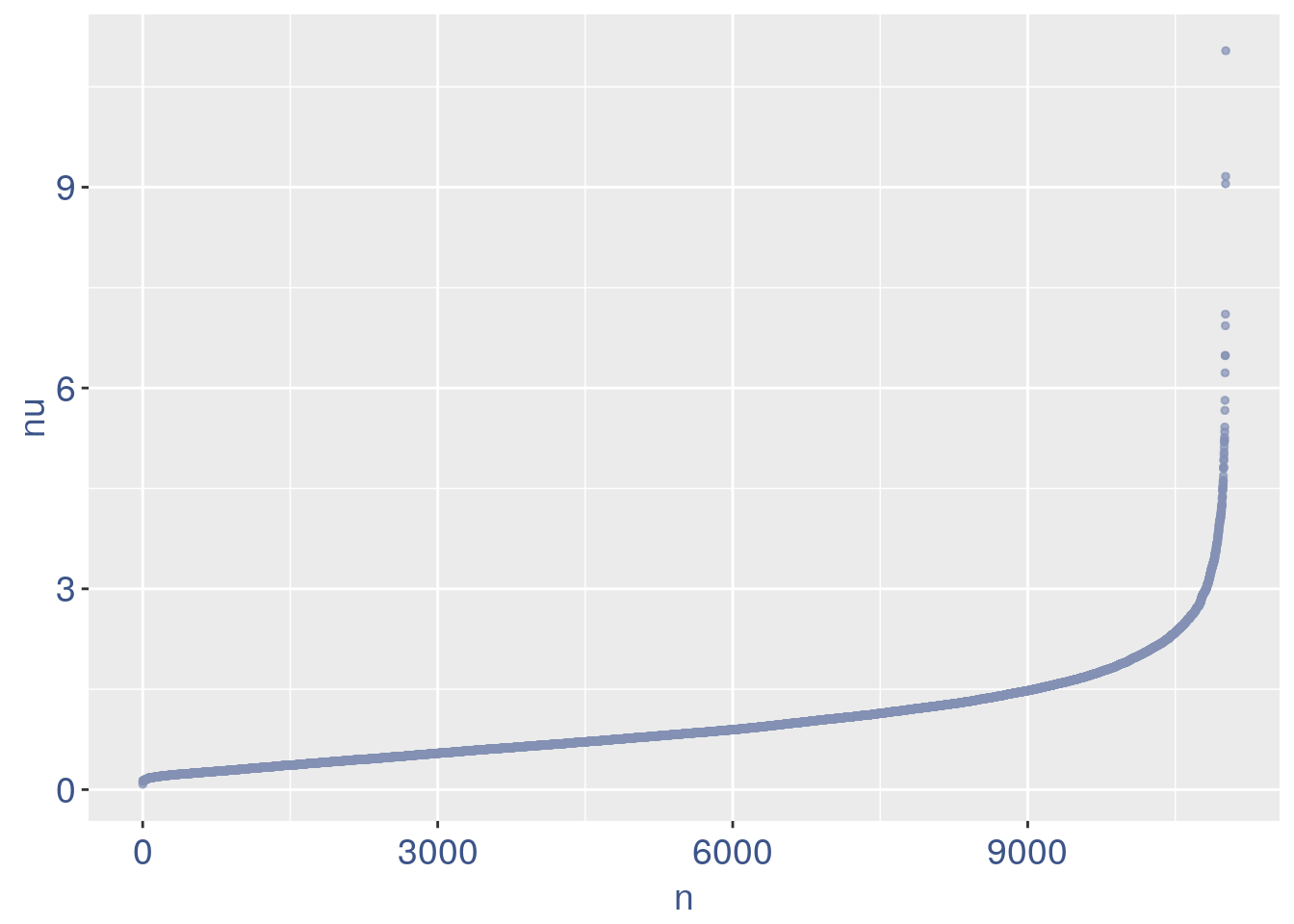
We can zoom on the smallest values and, if we detect a clear elbow, we can decide to remove the cells.
yset = 0.18 #threshold to remove low UDE cells
plot.ude <- ggplot(nu_df, aes(x = n, y = nu)) +
geom_point(colour = "#8491B4B2", size = 1) +
my_theme + ylim(0, 0.7) + xlim(0, 2000) +
geom_hline(yintercept = yset, linetype = "dashed",
color = "darkred") + annotate(geom = "text",
x = 500, y = 0.5, label = paste("to remove cells with nu < ",
yset, sep = " "), color = "darkred",
size = 4.5)
pdf(paste(out_dir, "cleaning/", t, "_plots_efficiency.pdf",
sep = ""))
plot.ude
dev.off()
#> png
#> 2
plot.ude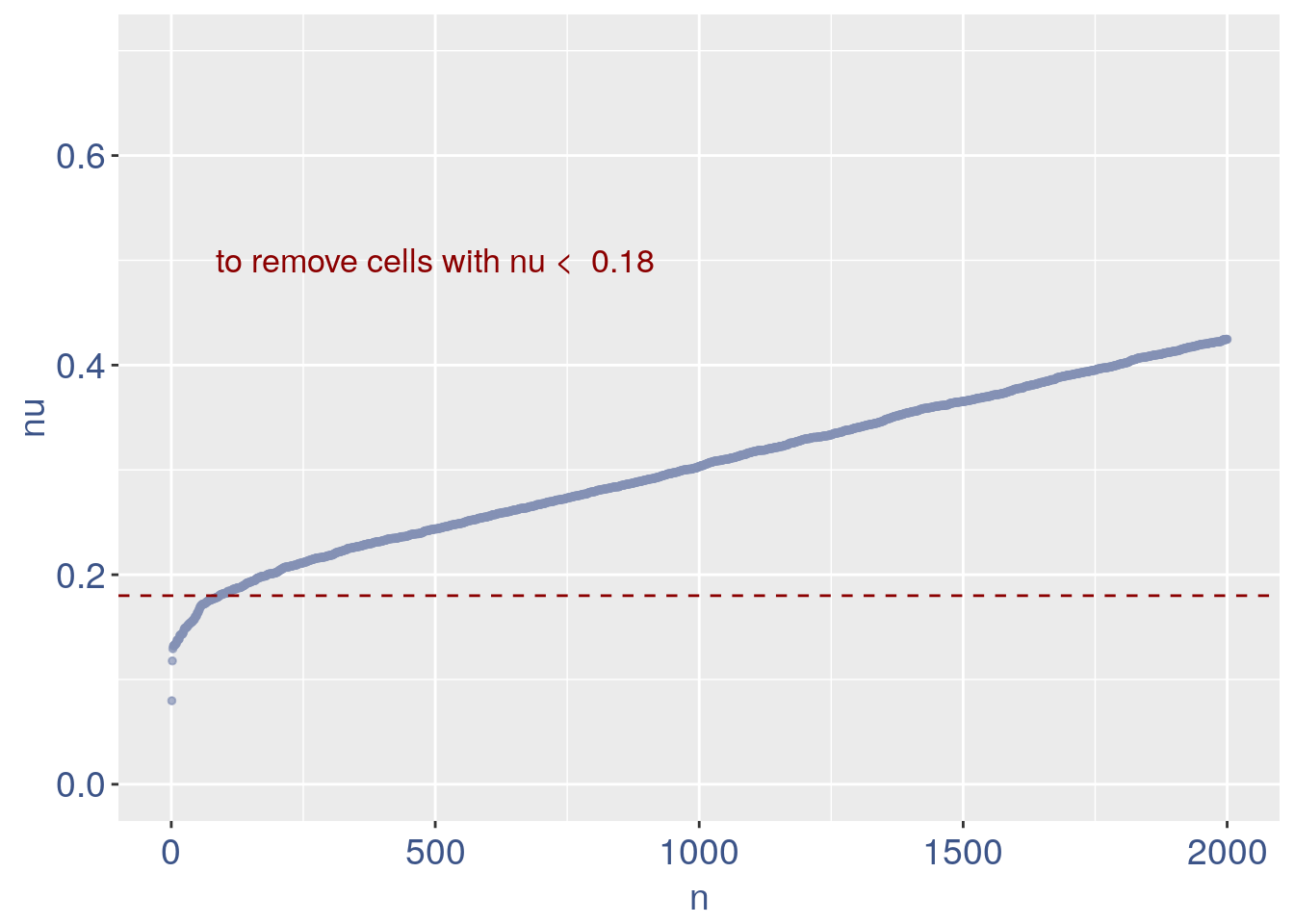
We also save the defined threshold in the metadata and re-run the estimation.
obj@meta[(nrow(obj@meta) + 1), 1:2] = c("Threshold low UDE cells:",
yset)
to_rem = rownames(nu_df[which(nu_df$nu <
yset), ])
obj@raw = obj@raw[, !colnames(obj@raw) %in%
to_rem]Repeat the estimation after the cells are removed.
ttm = clean(obj)
#> [1] "Start estimation mu with linear method"
#> [1] 13683 10923
#> rowname e14-WT10_AGAGCTGGACGT e14-WT10_CATGGTAAGACC
#> 7247 Meg3 84.952477 120.788082
#> 7016 Malat1 62.596562 120.788082
#> 11116 Snhg11 40.240647 65.645697
#> 7043 Map1b 0.000000 5.251656
#> 12527 Tuba1a 40.240647 28.884107
#> 12489 Ttc3 22.355915 0.000000
#> 8079 Nnat 4.471183 18.380795
#> 5606 Gria2 0.000000 13.129139
#> 5957 Hnrnpa2b1 17.884732 7.877484
#> 7047 Map2 8.942366 2.625828
#> 6024 Hsp90ab1 8.942366 2.625828
#> 7812 Nav1 4.471183 2.625828
#> 6513 Kif1b 8.942366 2.625828
#> 6533 Kif5c 0.000000 2.625828
#> 10318 Rtn1 22.355915 7.877484
#> e14-WT10_TACATACAATGG e14-WT10_TACGACGAATTA e14-WT8-1_ACCCACACTATC
#> 7247 121.310609 132.590823 140.839206
#> 7016 103.470813 185.627152 111.700060
#> 11116 0.000000 13.259082 29.139146
#> 7043 7.135918 26.518165 4.856524
#> 12527 24.975714 26.518165 4.856524
#> 12489 3.567959 6.629541 14.569573
#> 8079 7.135918 16.573853 0.000000
#> 5606 7.135918 13.259082 29.139146
#> 5957 7.135918 3.314771 4.856524
#> 7047 3.567959 3.314771 4.856524
#> 6024 0.000000 6.629541 4.856524
#> 7812 10.703877 13.259082 14.569573
#> 6513 0.000000 13.259082 4.856524
#> 6533 7.135918 13.259082 4.856524
#> 10318 17.839795 6.629541 4.856524
#> e14-WT8-1_GACACGGGACCC e14-WT8-1_GGCTACTGTGGT e14-WT8-1_TACATGATCGAT
#> 7247 91.647101 104.062121 212.756855
#> 7016 53.058848 80.047786 29.201921
#> 11116 0.000000 4.002389 58.403843
#> 7043 4.823532 16.009557 83.434061
#> 12527 24.117658 8.004779 16.686812
#> 12489 9.647063 16.009557 16.686812
#> 8079 14.470595 4.002389 0.000000
#> 5606 14.470595 4.002389 8.343406
#> 5957 14.470595 12.007168 4.171703
#> 7047 0.000000 12.007168 16.686812
#> 6024 4.823532 28.016725 12.515109
#> 7812 0.000000 12.007168 12.515109
#> 6513 14.470595 0.000000 8.343406
#> 6533 0.000000 20.011946 8.343406
#> 10318 0.000000 0.000000 8.343406
#> e14-WT8-2_AGTCTTCTTCAC e14-WT8-2_CCGACCTCATTG e14-WT8-2_GACCCTTTTCTA
#> 7247 111.760903 109.837945 141.225596
#> 7016 59.301704 168.679701 99.831887
#> 11116 0.000000 11.768351 19.479393
#> 7043 10.263756 7.845567 29.219089
#> 12527 38.774191 23.536702 12.174620
#> 12489 20.527513 11.768351 43.828633
#> 8079 5.702087 62.764540 9.739696
#> 5606 6.842504 11.768351 21.914317
#> 5957 4.561670 7.845567 7.304772
#> 7047 3.421252 7.845567 17.044468
#> 6024 11.404174 7.845567 2.434924
#> 7812 7.982922 0.000000 7.304772
#> 6513 5.702087 3.922784 14.609544
#> 6533 2.280835 3.922784 9.739696
#> 10318 6.842504 3.922784 0.000000
#> e14-WT8-2_TAACCACCCCGT e14-WT8-2_TACGTGGCGTCC e14-WT8-2_TCCTAGCCTGGT
#> 7247 112.37788 128.627510 205.079707
#> 7016 102.16171 107.189592 89.614662
#> 11116 20.43234 4.287584 46.530690
#> 7043 10.21617 51.451004 8.616794
#> 12527 15.32426 8.575167 5.170077
#> 12489 0.00000 21.437918 25.850383
#> 8079 10.21617 30.013086 5.170077
#> 5606 15.32426 8.575167 3.446718
#> 5957 25.54043 8.575167 3.446718
#> 7047 0.00000 21.437918 17.233589
#> 6024 0.00000 4.287584 6.893436
#> 7812 10.21617 12.862751 3.446718
#> 6513 20.43234 0.000000 13.786871
#> 6533 0.00000 8.575167 12.063512
#> 10318 15.32426 8.575167 6.893436
#> e14-WT9-2_TGGTTCTGCACT
#> 7247 95.068229
#> 7016 40.743527
#> 11116 21.729881
#> 7043 62.473408
#> 12527 27.162351
#> 12489 16.297411
#> 8079 10.864940
#> 5606 8.148705
#> 5957 10.864940
#> 7047 5.432470
#> 6024 21.729881
#> 7812 10.864940
#> 6513 10.864940
#> 6533 24.446116
#> 10318 2.716235
obj = ttm$object
ttm$pca.cell.2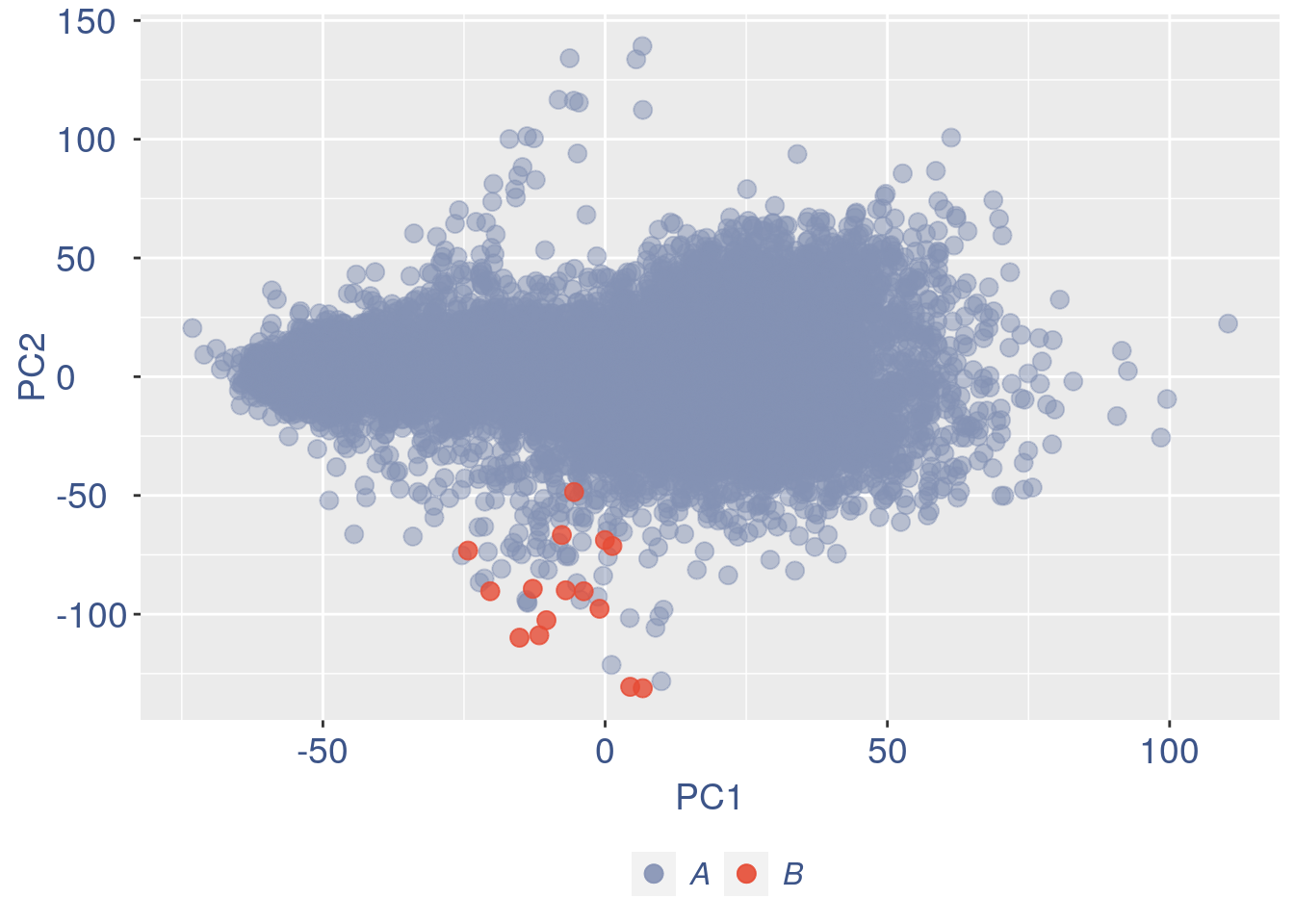
Just to check again, we plot the final efficiency coloured PCA.
nu_est = round(obj@nu, digits = 7)
plot.nu <- ggplot(ttm$pca_cells, aes(x = PC1,
y = PC2, colour = log(nu_est)))
plot.nu = plot.nu + geom_point(size = 2,
alpha = 0.8) + scale_color_gradient2(low = "#E64B35B2",
mid = "#4DBBD5B2", high = "#3C5488B2",
midpoint = log(mean(nu_est)), name = "ln(nu)") +
ggtitle("Cells PCA coloured by cells efficiency: last") +
my_theme + theme(plot.title = element_text(color = "#3C5488FF",
size = 20), legend.title = element_text(color = "#3C5488FF",
size = 14, face = "italic"), legend.text = element_text(color = "#3C5488FF",
size = 11), legend.key.width = unit(2,
"mm"), legend.position = "right")
pdf(paste(out_dir, "cleaning/", t, "_plots_PCA_efficiency_colored_FINAL.pdf",
sep = ""))
plot.nu
dev.off()
#> png
#> 2
plot.nu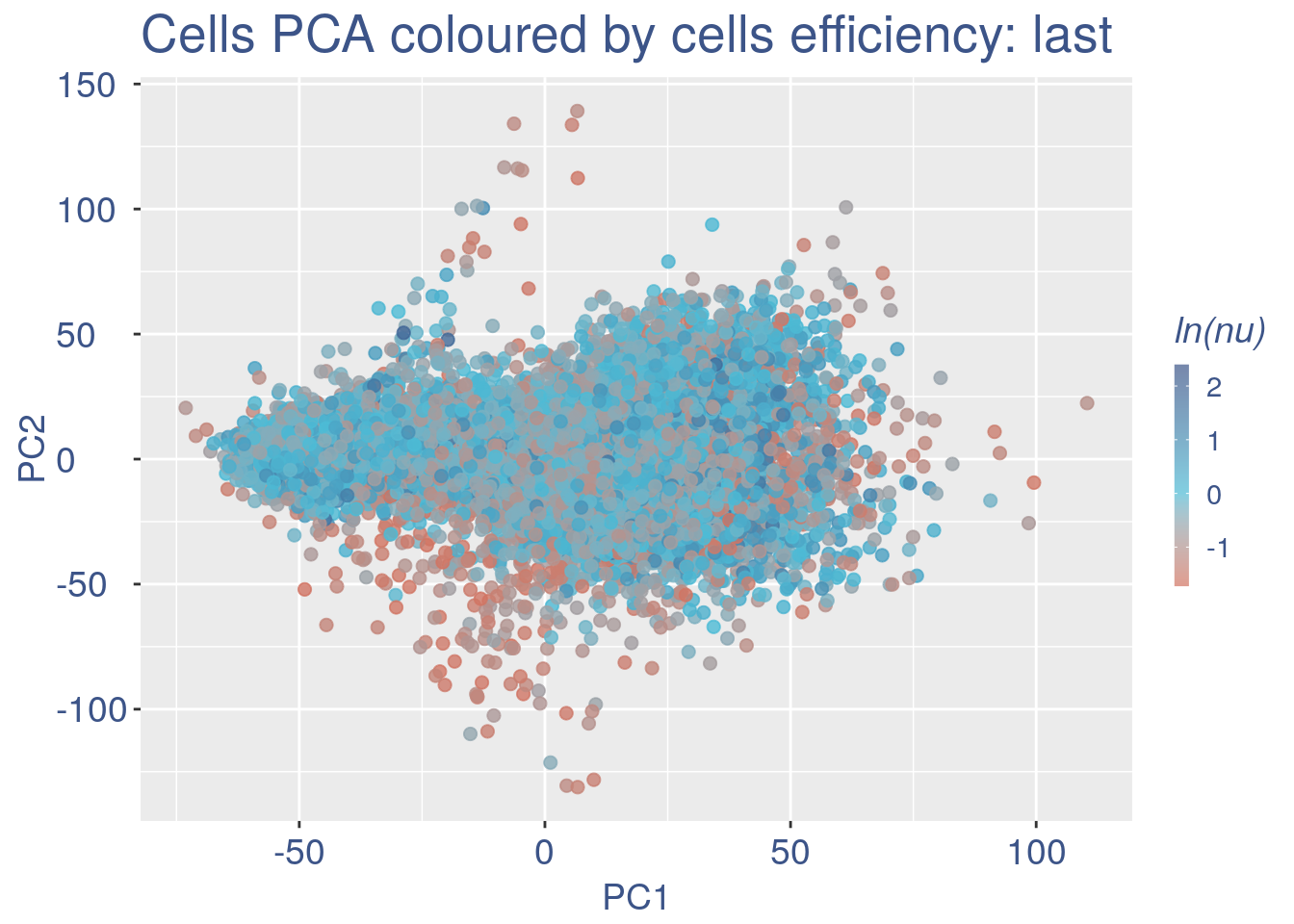
COTAN analysis: in this part all the contingency tables are computed and used to get the statistics (S) To storage efficiency of all the observed tables only the yes_yes is stored. Note that if will be necessary re-computing the yes-yes table, the yes-yes table should be cancelled before running cotan_analysis.
COEX evaluation and storing
0.3 Elaborated data analysis
0.3.1 GDI
The next function can directly plot the GDI for the dataset with the 1.5 threshold (in red) and the two higher quantiles (in blue).
obj = readRDS(paste(out_dir, t, ".cotan.RDS",
sep = ""))
plot_GDI(obj, cond = "E14.5")
#> [1] "GDI plot "
#> [1] "function to generate GDI dataframe"
#> [1] "Using S"
#> [1] "function to generate S "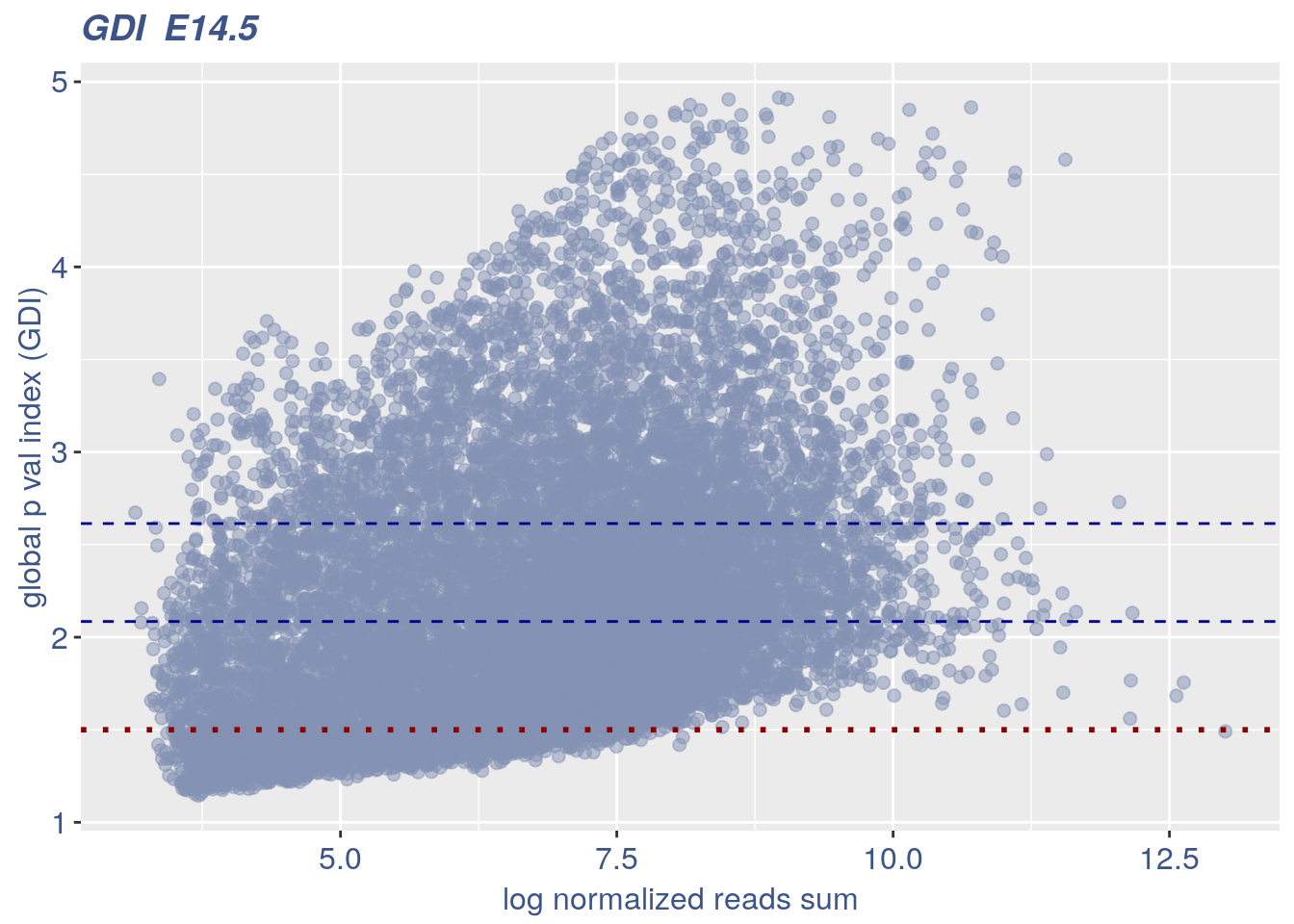
If a more complex plot is needed, or if we want to analyse mor in detail the GDI data, we can get directly the GDI data frame.
quant.p = get.GDI(obj)
#> [1] "function to generate GDI dataframe"
#> [1] "Using S"
#> [1] "function to generate S "
head(quant.p)
#> sum.raw.norm GDI exp.cells
#> 0610007N19Rik 7.210233 3.729326 8.239495
#> 0610007P14Rik 8.008533 2.254515 21.642406
#> 0610009B22Rik 8.030828 1.731632 21.697336
#> 0610009D07Rik 9.547315 2.298977 62.391284
#> 0610009E02Rik 5.742891 2.932436 2.728188
#> 0610009L18Rik 6.456446 1.492351 5.044402In the third column of this dataframe is reported the percentage of cells expressing the gene.
NPGs = c("Nes", "Vim", "Sox2", "Sox1", "Notch1",
"Hes1", "Hes5", "Pax6") #,'Hes3'
PNGs = c("Map2", "Tubb3", "Neurod1", "Nefm",
"Nefl", "Dcx", "Tbr1")
hk = c("Calm1", "Cox6b1", "Ppia", "Rpl18",
"Cox7c", "Erh", "H3f3a", "Taf1", "Taf2",
"Gapdh", "Actb", "Golph3", "Mtmr12",
"Zfr", "Sub1", "Tars", "Amacr")
text.size = 12
quant.p$highlight = with(quant.p, ifelse(rownames(quant.p) %in%
NPGs, "NPGs", ifelse(rownames(quant.p) %in%
hk, "Constitutive", ifelse(rownames(quant.p) %in%
PNGs, "PNGs", "normal"))))
textdf <- quant.p[rownames(quant.p) %in%
c(NPGs, hk, PNGs), ]
mycolours <- c(Constitutive = "#00A087FF",
NPGs = "#E64B35FF", PNGs = "#F39B7FFF",
normal = "#8491B4B2")
f1 = ggplot(subset(quant.p, highlight ==
"normal"), aes(x = sum.raw.norm, y = GDI)) +
geom_point(alpha = 0.1, color = "#8491B4B2",
size = 2.5)
GDI_plot = f1 + geom_point(data = subset(quant.p,
highlight != "normal"), aes(x = sum.raw.norm,
y = GDI, colour = highlight), size = 2.5,
alpha = 0.8) + geom_hline(yintercept = quantile(quant.p$GDI)[4],
linetype = "dashed", color = "darkblue") +
geom_hline(yintercept = quantile(quant.p$GDI)[3],
linetype = "dashed", color = "darkblue") +
geom_hline(yintercept = 1.5, linetype = "dotted",
color = "red", size = 0.5) + scale_color_manual("Status",
values = mycolours) + scale_fill_manual("Status",
values = mycolours) + xlab("log normalized counts") +
ylab("GDI") + geom_label_repel(data = textdf,
aes(x = sum.raw.norm, y = GDI, label = rownames(textdf),
fill = highlight), label.size = NA,
alpha = 0.5, direction = "both", na.rm = TRUE,
seed = 1234) + geom_label_repel(data = textdf,
aes(x = sum.raw.norm, y = GDI, label = rownames(textdf)),
label.size = NA, segment.color = "black",
segment.size = 0.5, direction = "both",
alpha = 0.8, na.rm = TRUE, fill = NA,
seed = 1234) + theme(axis.text.x = element_text(size = text.size,
angle = 0, hjust = 0.5, vjust = 0.5,
face = "plain", colour = "#3C5488FF"),
axis.text.y = element_text(size = text.size,
angle = 0, hjust = 0, vjust = 0.5,
face = "plain", colour = "#3C5488FF"),
axis.title.x = element_text(size = text.size,
angle = 0, hjust = 0.5, vjust = 0,
face = "plain", colour = "#3C5488FF"),
axis.title.y = element_text(size = text.size,
angle = 90, hjust = 0.5, vjust = 0.5,
face = "plain", colour = "#3C5488FF"),
legend.title = element_blank(), legend.text = element_text(color = "#3C5488FF",
face = "italic"), legend.position = "bottom") # titl)
legend <- cowplot::get_legend(GDI_plot)
GDI_plot = GDI_plot + theme(legend.position = "none")
GDI_plot
0.3.2 Heatmaps
For the Gene Pair Analysis, we can plot an heatmap with the coex values between two genes sets. To do so we need to define, in a list, the different gene sets (list.genes). Then in the function parameter sets we can decide which sets need to be considered (in the example from 1 to 3). In the condition parameter we should insert an array with each file name prefix to be considered (in the example, the file names is “E14_cortex”).
list.genes = list(Ref.col = PNGs, NPGs = NPGs,
Const. = hk)
plot_heatmap(df_genes = list.genes, sets = c(1:3),
conditions = "E14_cortex", dir = "Data/")
#> [1] "plot heatmap"
#> [1] "Loading condition E14_cortex"
#> [1] "Map2" "Tubb3" "Neurod1" "Nefm" "Nefl" "Dcx" "Tbr1"
#> [1] "Get p-values on a set of genes on columns on a set of genes on rows"
#> [1] "Using function S"
#> [1] "function to generate S "
#> [1] "Ref.col"
#> [1] "NPGs"
#> [1] "Const."
#> [1] "min coex: -0.00367088579639376 max coex 0.00265975905519931"
print(sessionInfo())
#> R version 4.0.4 (2021-02-15)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Ubuntu 18.04.5 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.2.20.so
#>
#> locale:
#> [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
#> [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
#> [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
#> [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
#> [9] LC_ADDRESS=C LC_TELEPHONE=C
#> [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] ggrepel_0.9.1 ggplot2_3.3.3 Matrix_1.3-2 data.table_1.14.0
#> [5] COTAN_0.1.0
#>
#> loaded via a namespace (and not attached):
#> [1] sass_0.3.1 tidyr_1.1.2 jsonlite_1.7.2
#> [4] R.utils_2.10.1 bslib_0.2.4 assertthat_0.2.1
#> [7] highr_0.8 stats4_4.0.4 yaml_2.2.1
#> [10] pillar_1.5.1 lattice_0.20-41 glue_1.4.2
#> [13] reticulate_1.18 digest_0.6.27 RColorBrewer_1.1-2
#> [16] colorspace_2.0-0 cowplot_1.1.1 htmltools_0.5.1.1
#> [19] R.oo_1.24.0 pkgconfig_2.0.3 GetoptLong_1.0.5
#> [22] purrr_0.3.4 scales_1.1.1 tibble_3.1.0
#> [25] generics_0.1.0 farver_2.1.0 IRanges_2.24.1
#> [28] ellipsis_0.3.1 withr_2.4.1 BiocGenerics_0.36.0
#> [31] magrittr_2.0.1 crayon_1.4.0 evaluate_0.14
#> [34] R.methodsS3_1.8.1 fansi_0.4.2 Cairo_1.5-12.2
#> [37] tools_4.0.4 GlobalOptions_0.1.2 formatR_1.8
#> [40] lifecycle_1.0.0 matrixStats_0.58.0 ComplexHeatmap_2.6.2
#> [43] basilisk.utils_1.2.2 stringr_1.4.0 S4Vectors_0.28.1
#> [46] munsell_0.5.0 cluster_2.1.1 compiler_4.0.4
#> [49] jquerylib_0.1.3 rlang_0.4.10 grid_4.0.4
#> [52] rjson_0.2.20 rappdirs_0.3.3 circlize_0.4.12
#> [55] labeling_0.4.2 rmarkdown_2.7 basilisk_1.2.1
#> [58] gtable_0.3.0 DBI_1.1.1 R6_2.5.0
#> [61] knitr_1.31 dplyr_1.0.4 utf8_1.2.1
#> [64] clue_0.3-58 filelock_1.0.2 shape_1.4.5
#> [67] stringi_1.5.3 parallel_4.0.4 Rcpp_1.0.6
#> [70] vctrs_0.3.6 png_0.1-7 tidyselect_1.1.0
#> [73] xfun_0.22