#library(SingleCellExperiment)
#library(DuoClustering2018)
#library(tidyr)
library(rlang)
library(ggplot2)
library(ggsankey) # remotes::install_github("davidsjoberg/ggsankey")
library(tibble)
library(zeallot)
library(assertthat)
library(COTAN)
library(caret)
theme_set(theme_bw())
library(stringr)
library(nnet)
#devtools::load_all("~/dev/COTAN/COTAN/")
options(parallelly.fork.enable = TRUE)
outDir <- "Results/Clusterization/"
setLoggingLevel(2)
setLoggingFile(file.path(outDir, "CD14_Monocytes_ClusterizationsComparisons.log"))CD14+ clusterizations comparisons with CellTypist
cd14Obj <- readRDS(file = file.path("Data/CD14Cleaned/", "CD14_Monocytes.cotan.RDS"))
sampleCondition <- getMetadataElement(cd14Obj, datasetTags()[["cond"]])
sampleCondition[1] "CD14_Monocytes"getClusterizations(cd14Obj)[1] "split" "merge" "majority-voting"metaC <- getMetadataCells(cd14Obj)splitClusters <- getClusters(cd14Obj, "split")
mergedClusters <- getClusters(cd14Obj, "merge")labelsDF <- read.csv(file.path("Data/CD14Cleaned/", "CD14Cleaned_Immune_All_Low_predicted_labels.csv"), header = TRUE)
labelsDF <- column_to_rownames(labelsDF, var = "X")
rownames(labelsDF) <- gsub("[.]", "-", rownames(labelsDF))
cells_to_keep <- rownames(labelsDF)[rownames(labelsDF) %in% getCells(cd14Obj)]
assert_that(identical(cells_to_keep, getCells(cd14Obj)))
majorityVotingClusters <- labelsDF[cells_to_keep, "majority_voting"]
names(majorityVotingClusters) <- cells_to_keep
majorityVotingCoexDF <- DEAOnClusters(cd14Obj,clName ="majority-voting",clusters = majorityVotingClusters)
cd14Obj <- addClusterization(cd14Obj, clName = "majority-voting",
clusters = majorityVotingClusters,
coexDF = majorityVotingCoexDF)Save the COTAN object
saveRDS(cd14Obj, file = file.path(outDir, paste0(sampleCondition, ".cotan.RDS")))nlevels(splitClusters)
head(sort(table(splitClusters), decreasing = TRUE), 10L)
nlevels(mergedClusters)
head(sort(table(mergedClusters), decreasing = TRUE), 10L)
nlevels(majorityVotingClusters)
head(sort(table(majorityVotingClusters), decreasing = TRUE), 10L)splitClustersDF <- as.data.frame(splitClusters)
splitClustersDF[["cell"]] <- rownames(splitClustersDF)
colnames(splitClustersDF)[[1]] <- "COTAN.split.cluster"
splitClustersDF <- splitClustersDF[order(splitClustersDF[["COTAN.split.cluster"]]), ]
mergedClustersDF <- as.data.frame(mergedClusters)
mergedClustersDF[["cell"]] <- rownames(mergedClustersDF)
colnames(mergedClustersDF)[[1]] <- "COTAN.merged.cluster"
mergedClustersDF <- mergedClustersDF[order(mergedClustersDF[["COTAN.merged.cluster"]]), ]
majorityVotingClustersDF <- as.data.frame(majorityVotingClusters)
majorityVotingClustersDF[["cell"]] <- rownames(majorityVotingClustersDF)
colnames(majorityVotingClustersDF)[[1]] <- "majority.voting.cluster"
majorityVotingClustersDF <- majorityVotingClustersDF[order(majorityVotingClustersDF[["majority.voting.cluster"]]), ]mjvt_split.table <- merge.data.frame(x = majorityVotingClustersDF, y = splitClustersDF,
by = "cell", all.x = TRUE, all.y = TRUE)
table(mjvt_split.table[,c(2L, 3L)]) COTAN.split.cluster
majority.voting.cluster 1 2 3 4 5
Classical monocytes 877 3 637 837 48
NK cells 1 25 0 0 0
pDC 0 10 0 0 0mjvt_split.table2 <- mjvt_split.table %>% make_long(majority.voting.cluster, COTAN.split.cluster)
ggplot(mjvt_split.table2,
aes(x = x,
next_x = next_x,
node = node,
next_node = next_node,
fill = factor(node),
label = node)) +
geom_sankey(flow.alpha = 0.75, node.color = 1) +
geom_sankey_label(size = 3.5, color = 1, fill = "white") +
scale_fill_viridis_d(option = "A", alpha = 0.95) +
theme_sankey(base_size = 16) +
theme(legend.position = "none")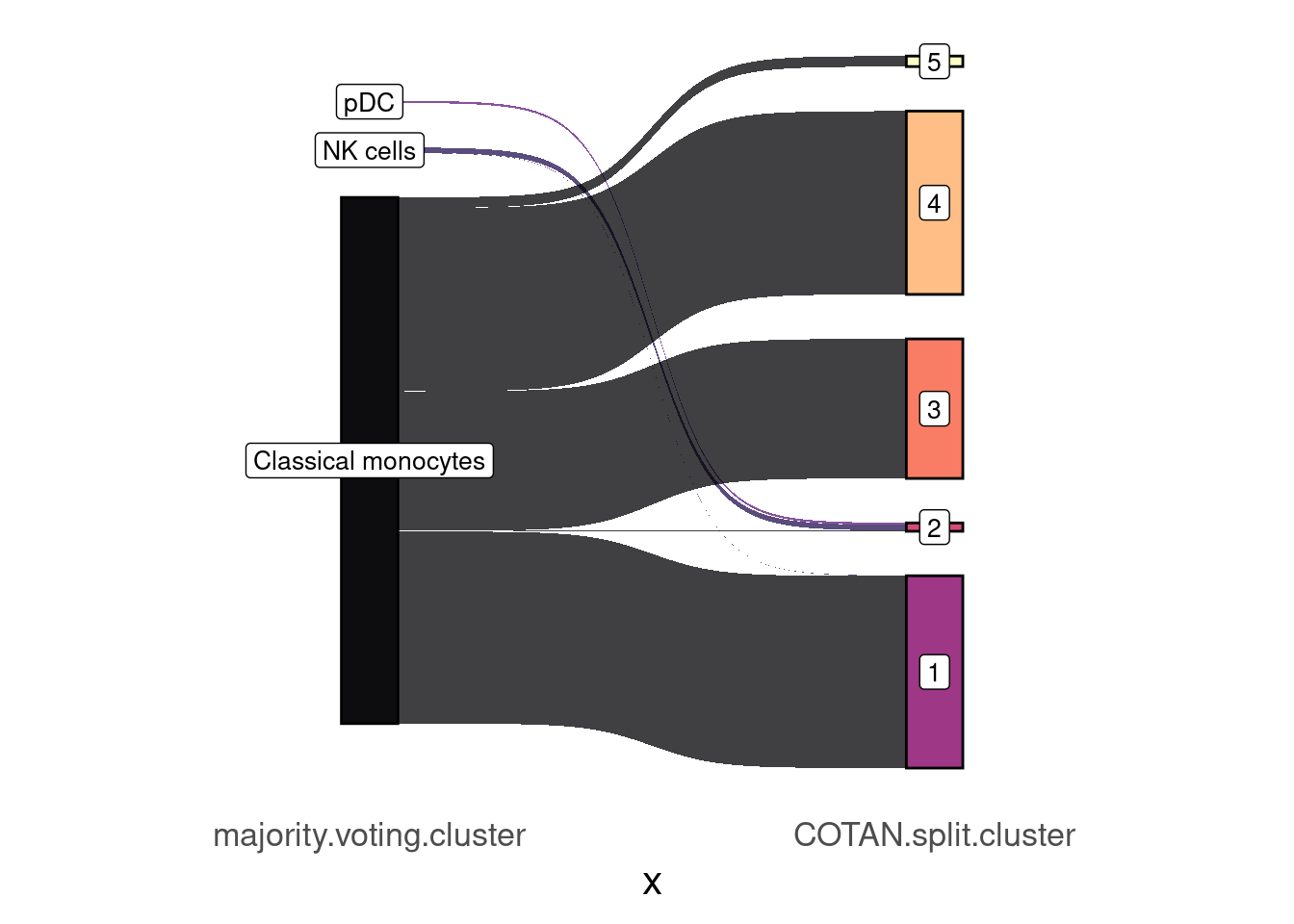
mjvt_merged.table <- merge.data.frame(x = majorityVotingClustersDF, y = mergedClustersDF,
by = "cell", all.x = TRUE, all.y = TRUE)
table(mjvt_merged.table[,c(2L, 3L)]) COTAN.merged.cluster
majority.voting.cluster 1 2 3
Classical monocytes 1522 877 3
NK cells 0 1 25
pDC 0 0 10mjvt_merged.table2 <- mjvt_merged.table %>% make_long(majority.voting.cluster, COTAN.merged.cluster)
ggplot(mjvt_merged.table2,
aes(x = x,
next_x = next_x,
node = node,
next_node = next_node,
fill = factor(node),
label = node)) +
geom_sankey(flow.alpha = 0.75, node.color = 1) +
geom_sankey_label(size = 3.5, color = 1, fill = "white") +
scale_fill_viridis_d(option = "A", alpha = 0.95) +
theme_sankey(base_size = 16) +
theme(legend.position = "none")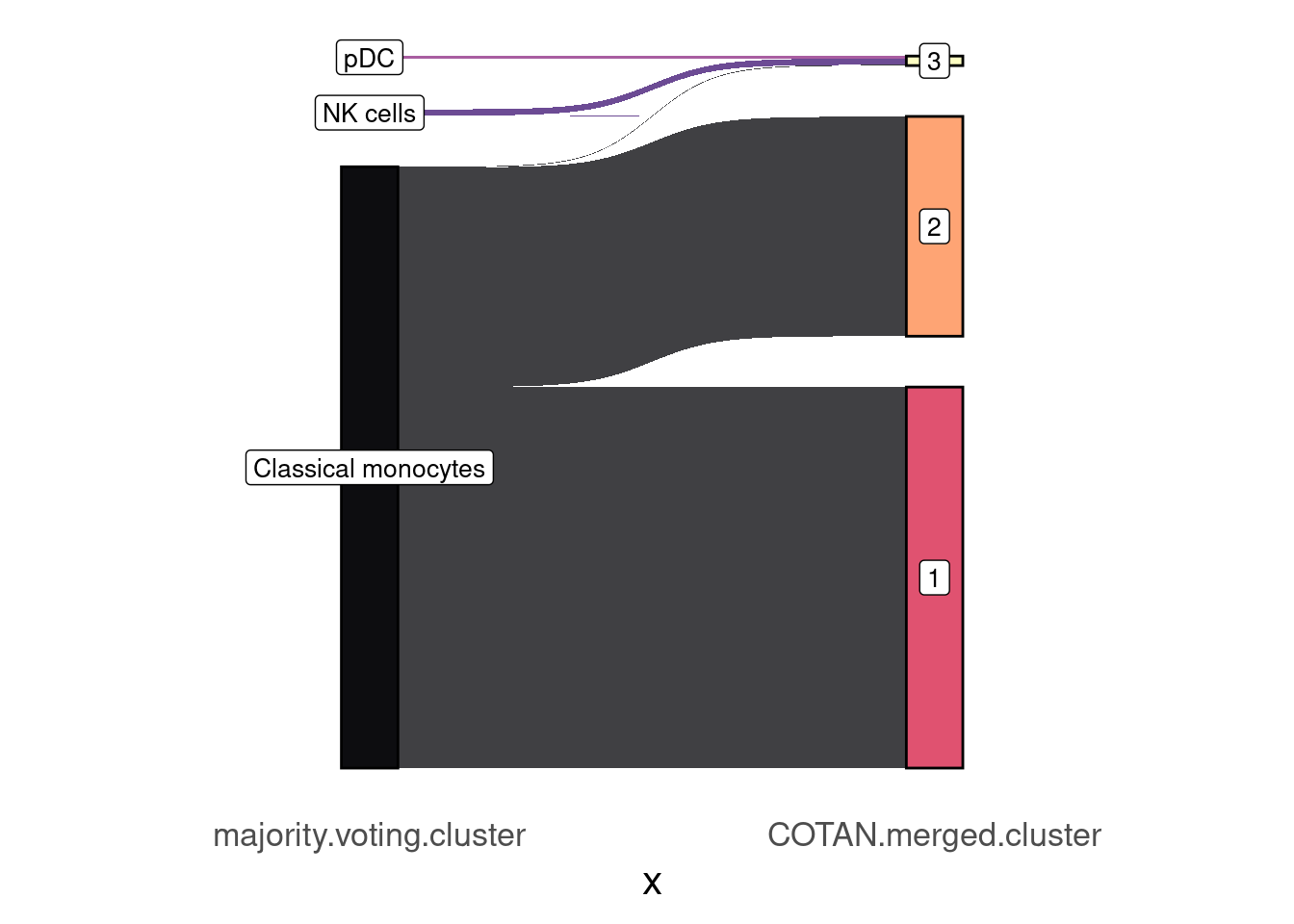
markersCD14 <- findClustersMarkers(cd14Obj,n = 100,clName = "merge",method = "BH")
write.csv(markersCD14,file = "Data/CD14Cleaned/ClusterMarkerGenes.csv")
head(markersCD14) CL Gene Score adjPVal DEA IsMarker logFoldCh
1 1 HLA-DQA1 -0.3398915 2.603676e-59 -0.15274999 0 -0.5470222
2 1 HLA-DQA2 -0.2899185 7.002901e-43 -0.12223685 0 -0.4804835
3 1 HLA-DPA1 -0.2808830 2.589504e-40 -0.13090249 0 -0.4370669
4 1 HLA-DRA -0.2499997 1.042557e-31 -0.10296185 0 -0.3654458
5 1 HLA-DPB1 -0.2225778 5.652934e-25 -0.09887797 0 -0.3375752
6 1 YBX1 -0.2095002 5.042102e-22 -0.08080263 0 -0.3784618cd14GDI <- calculateGDI(cd14Obj)
subsetGDI <- cd14GDI[cd14GDI$sum.raw.norm > 7,]
top.GDI.genes <- rownames(subsetGDI[order(subsetGDI$GDI,decreasing = T),])[1:50]
GDIPlot(cd14Obj,genes = "",GDIIn = cd14GDI)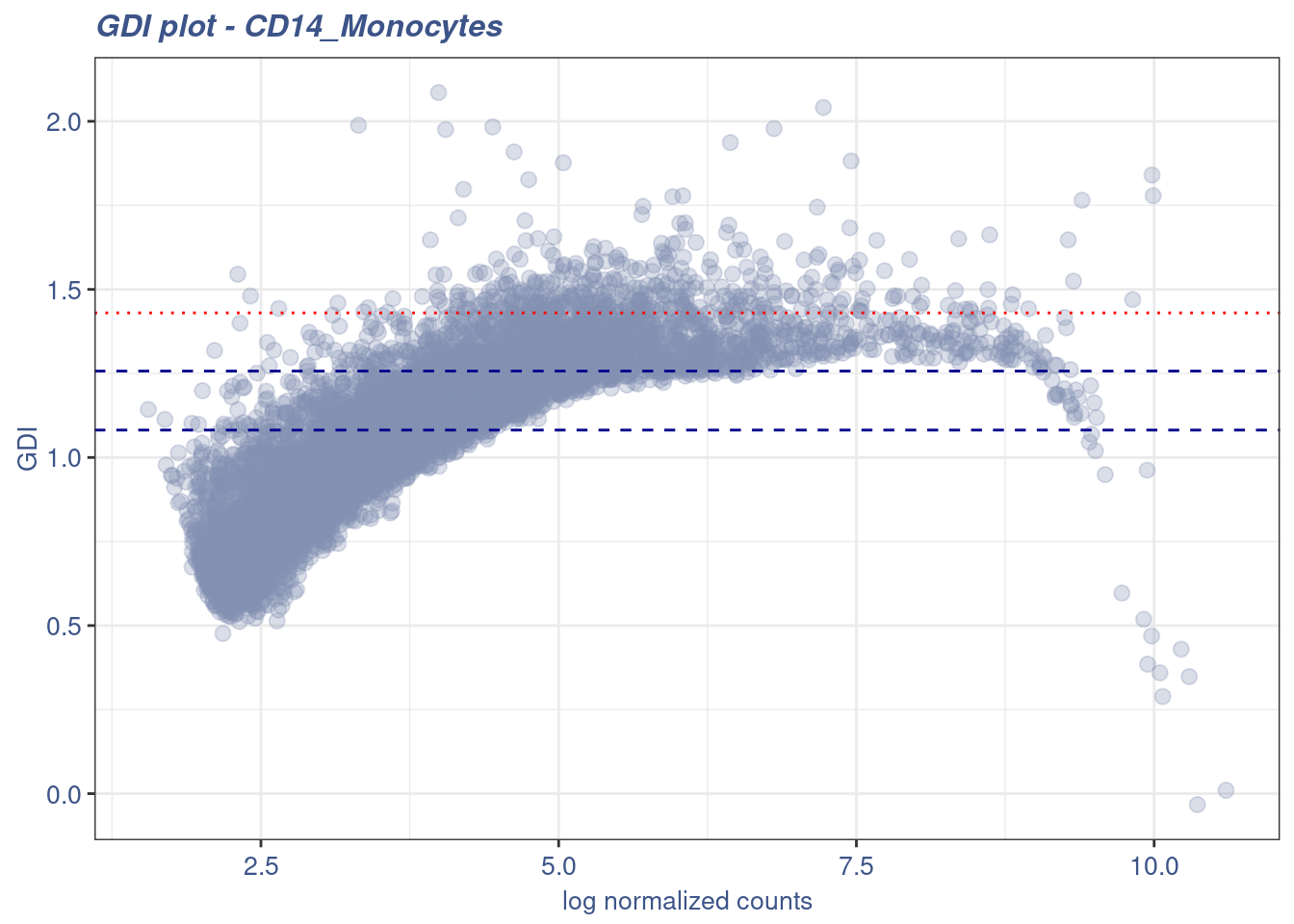
data <- getNormalizedData(cd14Obj)
data <- data[!rowSums(as.matrix(data)) < 1,]
data <- log(data*10000+1)
row_stdev <- apply(data, 1, sd, na.rm=TRUE)
row_stdev <- row_stdev[order(row_stdev,decreasing = T)]
genes.to.keep <- c(names(row_stdev[1:100]),top.GDI.genes)
data.small <- data[rownames(data) %in% genes.to.keep,]
#data <- t(as.matrix(data))
data.small <- t(as.matrix(data.small))
COTAN_Cl.code <- getClusterizationData(cd14Obj,clName = "merge")[[1]]
#COTAN_Cl.code[COTAN_Cl.code == "A549"] <- 0
#COTAN_Cl.code[COTAN_Cl.code == "CCL-185-IG"] <- 1
COTAN_Cl.code <- as.numeric(COTAN_Cl.code)
data.small <- cbind(data.small,COTAN_Cl.code)
data.small <- as.data.frame(data.small)
# Split the data into training and test set
set.seed(123)
training.samples <- data.small[,"COTAN_Cl.code"] %>%
createDataPartition(p = 0.8, list = FALSE)
train.data <- data.small[training.samples, ]
test.data <- data.small[-training.samples, ]
head(train.data) RPL22 LAPTM5 ATP6V0B RPL5 MCL1 CTSS
AAACATACCACTAG-1 9.485384 9.485384 0 0.000000 0.000000 0.000000
AAACATACGTTCAG-1 0.000000 0.000000 0 0.000000 10.114628 0.000000
AAACATTGCTTCGC-1 0.000000 0.000000 0 0.000000 0.000000 0.000000
AAACATTGGGCAAG-1 9.322304 0.000000 0 8.629246 9.727739 0.000000
AAACGGCTACGGAG-1 0.000000 0.000000 0 10.582413 9.483852 10.870089
AAACGGCTAGTCAC-1 0.000000 9.124915 0 9.124915 0.000000 9.818008
S100A10 S100A11 S100A9 S100A8 S100A6 S100A4
AAACATACCACTAG-1 9.485384 0.000000 9.485384 9.485384 9.485384 11.56476
AAACATACGTTCAG-1 0.000000 10.114628 11.906354 11.367363 10.807755 11.36736
AAACATTGCTTCGC-1 9.414354 10.512912 11.493724 11.812175 11.206045 11.71687
AAACATTGGGCAAG-1 10.238541 9.322304 10.826311 11.519449 10.420856 11.33713
AAACGGCTACGGAG-1 10.176961 0.000000 12.191831 11.881678 10.176961 11.56323
AAACGGCTAGTCAC-1 0.000000 10.511128 12.120544 11.763871 10.916584 11.20426
VAMP8 COX5B NBEAL1 EEF1B2 ARPC2 RPL15
AAACATACCACTAG-1 10.583946 0.000000 9.485384 0 10.178493 11.094761
AAACATACGTTCAG-1 10.114628 0.000000 0.000000 0 0.000000 9.421522
AAACATTGCTTCGC-1 9.414354 0.000000 9.414354 0 9.414354 9.414354
AAACATTGGGCAAG-1 9.727739 8.629246 9.322304 0 8.629246 11.113989
AAACGGCTACGGAG-1 10.176961 10.176961 0.000000 0 9.483852 10.582413
AAACGGCTAGTCAC-1 9.818008 9.818008 9.124915 0 9.818008 10.916584
RPSA GPX1 RPL29 SERP1 BTF3 CD14
AAACATACCACTAG-1 0.000000 9.485384 0.000000 0.000000 0.000000 10.178493
AAACATACGTTCAG-1 9.421522 9.421522 0.000000 0.000000 10.114628 10.520080
AAACATTGCTTCGC-1 0.000000 9.414354 10.512912 0.000000 0.000000 0.000000
AAACATTGGGCAAG-1 10.931670 10.420856 10.015406 9.322304 0.000000 0.000000
AAACGGCTACGGAG-1 0.000000 11.429697 9.483852 0.000000 0.000000 9.483852
AAACGGCTAGTCAC-1 0.000000 10.223455 9.124915 9.124915 9.124915 0.000000
DUSP1 HIGD2A PRELID1 HLA-E AIF1 CLIC1
AAACATACCACTAG-1 0.000000 9.485384 0.000000 0.000000 9.485384 10.58395
AAACATACGTTCAG-1 9.421522 0.000000 0.000000 0.000000 9.421522 0.00000
AAACATTGCTTCGC-1 9.414354 9.414354 0.000000 0.000000 11.206045 0.00000
AAACATTGGGCAAG-1 9.322304 9.322304 9.727739 8.629246 9.727739 0.00000
AAACGGCTACGGAG-1 0.000000 0.000000 0.000000 0.000000 0.000000 0.00000
AAACGGCTAGTCAC-1 9.124915 0.000000 9.124915 10.511128 9.818008 0.00000
HLA-DRA HLA-DRB5 HLA-DRB1 HLA-DQA1 HLA-DQB1 HLA-DMA
AAACATACCACTAG-1 10.583946 10.871621 11.883210 0 10.178493 0.000000
AAACATACGTTCAG-1 9.421522 0.000000 9.421522 0 0.000000 0.000000
AAACATTGCTTCGC-1 0.000000 10.107460 10.512912 0 0.000000 9.414354
AAACATTGGGCAAG-1 10.575003 10.015406 10.575003 0 9.727739 0.000000
AAACGGCTACGGAG-1 0.000000 9.483852 9.483852 0 0.000000 0.000000
AAACGGCTAGTCAC-1 10.223455 0.000000 9.818008 0 9.124915 0.000000
HLA-DPA1 HLA-DPB1 RPS10 SF3B5 RNASET2 RAC1
AAACATACCACTAG-1 9.485384 9.485384 0.000000 0.000000 0.000000 10.178493
AAACATACGTTCAG-1 0.000000 0.000000 9.421522 0.000000 0.000000 0.000000
AAACATTGCTTCGC-1 0.000000 9.414354 0.000000 0.000000 0.000000 9.414354
AAACATTGGGCAAG-1 0.000000 11.113989 9.727739 9.727739 8.629246 0.000000
AAACGGCTACGGAG-1 0.000000 0.000000 0.000000 0.000000 9.483852 0.000000
AAACGGCTAGTCAC-1 0.000000 0.000000 9.124915 0.000000 0.000000 9.124915
TOMM7 PPIA CHCHD2 ARPC1B TMEM176B SLC25A6 TIMP1
AAACATACCACTAG-1 9.485384 0.000000 0 9.485384 0.000000 10.178493 0
AAACATACGTTCAG-1 10.114628 9.421522 0 10.114628 0.000000 10.114628 0
AAACATTGCTTCGC-1 10.107460 0.000000 0 0.000000 9.414354 10.512912 0
AAACATTGGGCAAG-1 8.629246 0.000000 0 8.629246 0.000000 8.629246 0
AAACGGCTACGGAG-1 0.000000 0.000000 0 10.176961 9.483852 0.000000 0
AAACGGCTAGTCAC-1 0.000000 0.000000 0 9.818008 9.124915 9.124915 0
CFP CEBPD PABPC1 EDF1 KLF6 VIM
AAACATACCACTAG-1 0.00000 0.000000 0.000000 9.485384 10.178493 10.178493
AAACATACGTTCAG-1 10.11463 0.000000 0.000000 0.000000 10.114628 9.421522
AAACATTGCTTCGC-1 0.00000 9.414354 0.000000 0.000000 9.414354 0.000000
AAACATTGGGCAAG-1 10.70853 8.629246 0.000000 9.322304 10.015406 9.727739
AAACGGCTACGGAG-1 0.00000 11.093229 0.000000 0.000000 10.176961 0.000000
AAACGGCTAGTCAC-1 0.00000 10.223455 9.818008 0.000000 0.000000 9.818008
SRGN TALDO1 LSP1 MS4A6A NEAT1 ATP5L TPI1
AAACATACCACTAG-1 9.485384 0.000000 9.485384 9.485384 0.000000 0.000000 0.000000
AAACATACGTTCAG-1 0.000000 9.421522 0.000000 0.000000 9.421522 0.000000 0.000000
AAACATTGCTTCGC-1 0.000000 0.000000 0.000000 0.000000 9.414354 9.414354 0.000000
AAACATTGGGCAAG-1 8.629246 0.000000 9.727739 0.000000 9.322304 0.000000 8.629246
AAACGGCTACGGAG-1 9.483852 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000
AAACGGCTAGTCAC-1 9.124915 9.124915 0.000000 9.818008 0.000000 0.000000 0.000000
ARHGDIB CD63 NACA LINC00936 ARPC3 ATP5EP2
AAACATACCACTAG-1 0.000000 0.000000 0.000000 0.000000 10.178493 0.000000
AAACATACGTTCAG-1 0.000000 9.421522 9.421522 10.114628 0.000000 9.421522
AAACATTGCTTCGC-1 9.414354 0.000000 0.000000 0.000000 9.414354 10.512912
AAACATTGGGCAAG-1 8.629246 8.629246 9.727739 8.629246 0.000000 8.629246
AAACGGCTACGGAG-1 0.000000 0.000000 10.176961 9.483852 9.483852 0.000000
AAACGGCTAGTCAC-1 9.124915 9.124915 0.000000 0.000000 9.818008 9.124915
TPT1 PSME1 PSME2 NFKBIA RPL36AL NPC2
AAACATACCACTAG-1 0.000000 0.000000 10.178493 0.000000 0.000000 9.485384
AAACATACGTTCAG-1 9.421522 9.421522 0.000000 0.000000 9.421522 0.000000
AAACATTGCTTCGC-1 9.414354 0.000000 0.000000 0.000000 0.000000 0.000000
AAACATTGGGCAAG-1 0.000000 8.629246 9.322304 9.322304 9.322304 0.000000
AAACGGCTACGGAG-1 0.000000 0.000000 9.483852 0.000000 10.176961 0.000000
AAACGGCTAGTCAC-1 9.818008 0.000000 9.818008 9.124915 9.124915 9.818008
CRIP1 ANXA2 RPL4 TCEB2 CORO1A PYCARD
AAACATACCACTAG-1 0.00000 0.000000 10.58395 0.000000 9.485384 10.871621
AAACATACGTTCAG-1 0.00000 9.421522 10.11463 9.421522 0.000000 0.000000
AAACATTGCTTCGC-1 0.00000 0.000000 0.00000 0.000000 9.414354 9.414354
AAACATTGGGCAAG-1 10.42086 9.322304 0.00000 0.000000 8.629246 9.727739
AAACGGCTACGGAG-1 0.00000 9.483852 0.00000 0.000000 0.000000 10.176961
AAACGGCTAGTCAC-1 0.00000 0.000000 0.00000 0.000000 9.124915 9.818008
COTL1 APRT GABARAP EIF4A1 UBB RPL23 GRN
AAACATACCACTAG-1 0.000000 0.000000 9.485384 0.000000 0 0.000000 0
AAACATACGTTCAG-1 0.000000 0.000000 9.421522 0.000000 0 9.421522 0
AAACATTGCTTCGC-1 0.000000 0.000000 0.000000 10.107460 0 9.414354 0
AAACATTGGGCAAG-1 0.000000 8.629246 8.629246 9.322304 0 10.015406 0
AAACGGCTACGGAG-1 0.000000 0.000000 10.176961 0.000000 0 0.000000 0
AAACGGCTAGTCAC-1 9.124915 0.000000 9.124915 0.000000 0 0.000000 0
RPL38 ACTG1 SCAND1 PSMA7 RPS21 CFD
AAACATACCACTAG-1 0.000000 0.000000 9.485384 0.00000 0.000000 9.485384
AAACATACGTTCAG-1 10.114628 0.000000 0.000000 0.00000 9.421522 0.000000
AAACATTGCTTCGC-1 0.000000 0.000000 0.000000 10.51291 10.107460 10.512912
AAACATTGGGCAAG-1 8.629246 9.322304 0.000000 10.01541 8.629246 0.000000
AAACGGCTACGGAG-1 0.000000 0.000000 10.176961 0.00000 9.483852 0.000000
AAACGGCTAGTCAC-1 0.000000 9.124915 9.818008 10.22345 9.818008 9.124915
GPX4 ATP5D UQCR11.1 NDUFA11 C19orf43 JUNB
AAACATACCACTAG-1 11.88321 0.000000 0.000000 0.000000 9.485384 0.000000
AAACATACGTTCAG-1 0.00000 0.000000 10.114628 9.421522 0.000000 0.000000
AAACATTGCTTCGC-1 0.00000 0.000000 0.000000 0.000000 0.000000 9.414354
AAACATTGGGCAAG-1 0.00000 8.629246 9.727739 9.322304 8.629246 0.000000
AAACGGCTACGGAG-1 0.00000 10.176961 0.000000 9.483852 0.000000 0.000000
AAACGGCTAGTCAC-1 0.00000 0.000000 0.000000 9.818008 0.000000 0.000000
IER2 PGLS JUND NDUFA13 FXYD5 COX6B1
AAACATACCACTAG-1 0.000000 0.000000 9.485384 0.000000 0.000000 9.485384
AAACATACGTTCAG-1 11.030895 0.000000 0.000000 10.114628 0.000000 9.421522
AAACATTGCTTCGC-1 9.414354 0.000000 9.414354 10.107460 10.107460 0.000000
AAACATTGGGCAAG-1 0.000000 8.629246 8.629246 0.000000 9.322304 8.629246
AAACGGCTACGGAG-1 0.000000 0.000000 10.176961 9.483852 0.000000 9.483852
AAACGGCTAGTCAC-1 11.427402 0.000000 9.124915 9.124915 0.000000 9.818008
HCST TYROBP GMFG ZFP36 BLVRB FOSB
AAACATACCACTAG-1 0.000000 10.178493 0.000000 0.000000 0.000000 0.000000
AAACATACGTTCAG-1 0.000000 10.807755 10.114628 0.000000 0.000000 9.421522
AAACATTGCTTCGC-1 0.000000 10.800587 9.414354 0.000000 0.000000 9.414354
AAACATTGGGCAAG-1 8.629246 10.575003 9.322304 0.000000 9.322304 0.000000
AAACGGCTACGGAG-1 0.000000 9.483852 0.000000 0.000000 9.483852 0.000000
AAACGGCTAGTCAC-1 0.000000 11.522711 9.124915 9.818008 9.124915 9.818008
AP2S1 GLTSCR2 EMP3 FCGRT RPS5 LGALS2
AAACATACCACTAG-1 0.000000 9.485384 9.485384 0.000000 9.485384 10.178493
AAACATACGTTCAG-1 0.000000 9.421522 0.000000 0.000000 0.000000 0.000000
AAACATTGCTTCGC-1 0.000000 0.000000 9.414354 0.000000 10.107460 0.000000
AAACATTGGGCAAG-1 8.629246 0.000000 10.826311 0.000000 10.015406 9.727739
AAACGGCTACGGAG-1 10.176961 0.000000 10.870089 10.176961 10.176961 0.000000
AAACGGCTAGTCAC-1 0.000000 10.223455 0.000000 9.124915 0.000000 0.000000
MT-ND2 MT-ATP6 MT-ND4 MT-CYB COTAN_Cl.code
AAACATACCACTAG-1 0.000000 0.000000 9.485384 0.000000 1
AAACATACGTTCAG-1 0.000000 9.421522 0.000000 9.421522 1
AAACATTGCTTCGC-1 0.000000 9.414354 0.000000 9.414354 1
AAACATTGGGCAAG-1 8.629246 9.727739 0.000000 8.629246 2
AAACGGCTACGGAG-1 0.000000 0.000000 0.000000 9.483852 1
AAACGGCTAGTCAC-1 9.124915 9.124915 0.000000 0.000000 1train.data$COTAN_Cl.code <- as.factor(train.data$COTAN_Cl.code)
test.data$COTAN_Cl.code <- as.factor(test.data$COTAN_Cl.code)
# Fit the model
#model <- glm( COTAN_Cl.code ~., data = train.data, family = binomial,control = list(maxit = 75))
model <- multinom(COTAN_Cl.code ~ ., data = train.data, maxit = 500)# weights: 369 (244 variable)
initial value 2143.392575
iter 10 value 761.340646
iter 20 value 746.268326
iter 30 value 720.679315
iter 40 value 579.207155
iter 50 value 545.792232
iter 60 value 539.711792
iter 70 value 526.902032
iter 80 value 506.380523
iter 90 value 485.108188
iter 100 value 480.679083
iter 110 value 476.395523
iter 120 value 462.778421
iter 130 value 457.035815
iter 140 value 455.285417
iter 150 value 449.873877
iter 160 value 446.861152
iter 170 value 444.452737
iter 180 value 439.972413
iter 190 value 437.990151
iter 200 value 437.057149
iter 210 value 436.908590
iter 220 value 436.792687
iter 230 value 436.667356
iter 240 value 436.372853
iter 250 value 435.784259
iter 260 value 435.132550
iter 270 value 434.964836
iter 280 value 434.951070
iter 290 value 434.949618
iter 300 value 434.949374
final value 434.949335
convergedprobabilities <- predict(model, newdata = test.data, type = "probs")
# Find the class with the highest probability for each case
predicted.classes <- apply(probabilities, 1, which.max)
# Adjust predicted classes to match your factor levels
levels <- levels(train.data$COTAN_Cl.code)
predicted.classes <- levels[predicted.classes]
# Calculate model accuracy
accuracy <- mean(predicted.classes == test.data$COTAN_Cl.code)
#result.df[nrow(result.df),"Accuracy"] <- accuracy
print(accuracy)[1] 0.8521561So the logistic regression accuracy for COTAN three clusters is quite good.
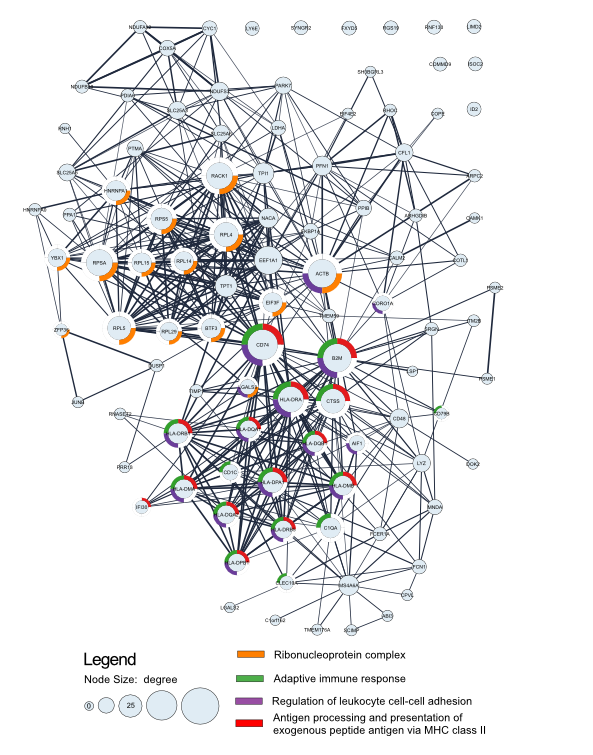
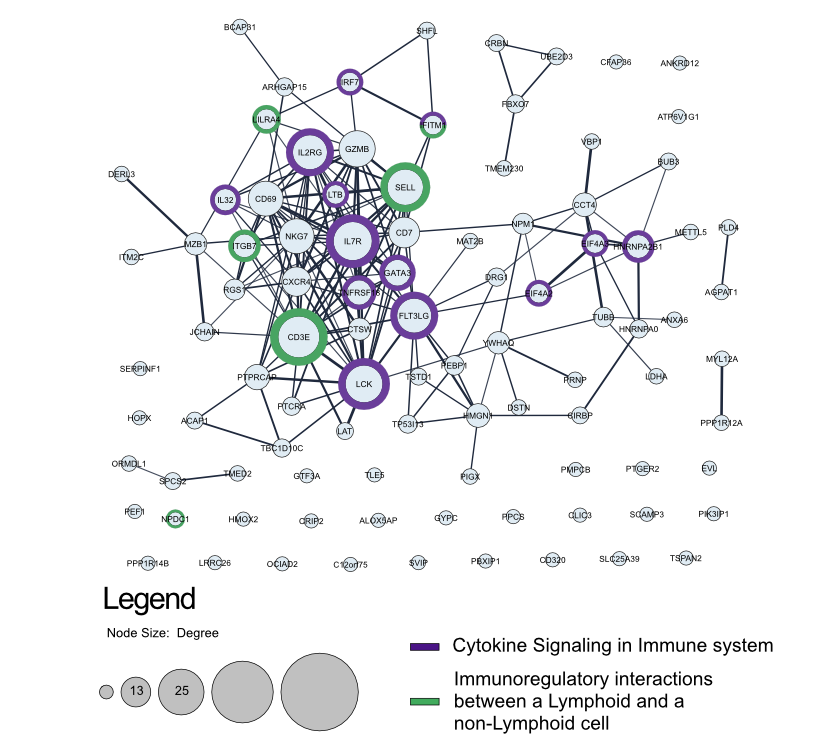
The cells in cluster 1 seem to be classical CD14 monocytes since they express CD14 while depleted in MHC class II proteins complex which is enriched in cell cluster 2 that seems intermediate monocytes.
Cluster 3 does not seem to be a monocyte cluster. Using enrichr website it is enriched in Plasmacytoid Dendritic cell marker genes.
sessionInfo()R version 4.4.0 (2024-04-24)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 20.04.6 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/liblapack.so.3; LAPACK version 3.9.0
locale:
[1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
[4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
[7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
time zone: Europe/Rome
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] nnet_7.3-19 stringr_1.5.1 caret_6.0-94 lattice_0.22-6
[5] COTAN_2.5.2 assertthat_0.2.1 zeallot_0.1.0 tibble_3.2.1
[9] ggsankey_0.0.99999 ggplot2_3.5.1 rlang_1.1.3
loaded via a namespace (and not attached):
[1] RcppAnnoy_0.0.22 splines_4.4.0
[3] later_1.3.2 polyclip_1.10-6
[5] hardhat_1.3.1 pROC_1.18.5
[7] rpart_4.1.23 fastDummies_1.7.3
[9] lifecycle_1.0.4 doParallel_1.0.17
[11] globals_0.16.3 MASS_7.3-60.2
[13] dendextend_1.17.1 magrittr_2.0.3
[15] plotly_4.10.4 rmarkdown_2.27
[17] yaml_2.3.8 httpuv_1.6.15
[19] Seurat_5.1.0 sctransform_0.4.1
[21] spam_2.10-0 askpass_1.2.0
[23] sp_2.1-4 spatstat.sparse_3.0-3
[25] reticulate_1.37.0 cowplot_1.1.3
[27] pbapply_1.7-2 RColorBrewer_1.1-3
[29] lubridate_1.9.3 abind_1.4-5
[31] zlibbioc_1.50.0 Rtsne_0.17
[33] purrr_1.0.2 BiocGenerics_0.50.0
[35] ipred_0.9-14 lava_1.8.0
[37] circlize_0.4.16 IRanges_2.38.0
[39] S4Vectors_0.42.0 ggrepel_0.9.5
[41] irlba_2.3.5.1 listenv_0.9.1
[43] spatstat.utils_3.0-4 umap_0.2.10.0
[45] goftest_1.2-3 RSpectra_0.16-1
[47] spatstat.random_3.2-3 dqrng_0.4.0
[49] fitdistrplus_1.1-11 parallelly_1.37.1
[51] DelayedMatrixStats_1.26.0 leiden_0.4.3.1
[53] codetools_0.2-20 DelayedArray_0.30.1
[55] tidyselect_1.2.1 shape_1.4.6.1
[57] farver_2.1.2 ScaledMatrix_1.12.0
[59] viridis_0.6.5 matrixStats_1.3.0
[61] stats4_4.4.0 spatstat.explore_3.2-7
[63] jsonlite_1.8.8 GetoptLong_1.0.5
[65] progressr_0.14.0 ggridges_0.5.6
[67] survival_3.6-4 iterators_1.0.14
[69] foreach_1.5.2 tools_4.4.0
[71] ica_1.0-3 Rcpp_1.0.12
[73] glue_1.7.0 prodlim_2023.08.28
[75] gridExtra_2.3 SparseArray_1.4.5
[77] xfun_0.44 MatrixGenerics_1.16.0
[79] ggthemes_5.1.0 dplyr_1.1.4
[81] withr_3.0.0 fastmap_1.2.0
[83] fansi_1.0.6 openssl_2.2.0
[85] digest_0.6.35 rsvd_1.0.5
[87] timechange_0.3.0 parallelDist_0.2.6
[89] R6_2.5.1 mime_0.12
[91] colorspace_2.1-0 scattermore_1.2
[93] tensor_1.5 spatstat.data_3.0-4
[95] utf8_1.2.4 tidyr_1.3.1
[97] generics_0.1.3 data.table_1.15.4
[99] recipes_1.0.10 class_7.3-22
[101] httr_1.4.7 htmlwidgets_1.6.4
[103] S4Arrays_1.4.1 ModelMetrics_1.2.2.2
[105] uwot_0.2.2 pkgconfig_2.0.3
[107] gtable_0.3.5 timeDate_4032.109
[109] ComplexHeatmap_2.20.0 lmtest_0.9-40
[111] XVector_0.44.0 htmltools_0.5.8.1
[113] dotCall64_1.1-1 clue_0.3-65
[115] SeuratObject_5.0.2 scales_1.3.0
[117] png_0.1-8 gower_1.0.1
[119] knitr_1.46 rstudioapi_0.16.0
[121] reshape2_1.4.4 rjson_0.2.21
[123] nlme_3.1-164 zoo_1.8-12
[125] GlobalOptions_0.1.2 KernSmooth_2.23-24
[127] parallel_4.4.0 miniUI_0.1.1.1
[129] RcppZiggurat_0.1.6 pillar_1.9.0
[131] grid_4.4.0 vctrs_0.6.5
[133] RANN_2.6.1 promises_1.3.0
[135] BiocSingular_1.20.0 beachmat_2.20.0
[137] xtable_1.8-4 cluster_2.1.6
[139] evaluate_0.23 cli_3.6.2
[141] compiler_4.4.0 crayon_1.5.2
[143] future.apply_1.11.2 labeling_0.4.3
[145] plyr_1.8.9 stringi_1.8.4
[147] viridisLite_0.4.2 deldir_2.0-4
[149] BiocParallel_1.38.0 munsell_0.5.1
[151] lazyeval_0.2.2 spatstat.geom_3.2-9
[153] PCAtools_2.16.0 Matrix_1.7-0
[155] RcppHNSW_0.6.0 patchwork_1.2.0
[157] sparseMatrixStats_1.16.0 future_1.33.2
[159] shiny_1.8.1.1 ROCR_1.0-11
[161] Rfast_2.1.0 igraph_2.0.3
[163] RcppParallel_5.1.7