import pandas as pd
import numpy as np
import scanpy as sc
from sklearn.metrics.cluster import normalized_mutual_info_score, adjusted_rand_score
from sklearn.metrics import homogeneity_score, completeness_score, fowlkes_mallows_score, silhouette_score, davies_bouldin_score
from sklearn.metrics.cluster import contingency_matrix
from src.utils import sankey_plot
import kaleido
from sklearn.preprocessing import StandardScaler
import plotly.io as pio
import matplotlib.pyplot as plt
import seaborn as snsClustering Comparision
Preamble
DIR = 'Data/'
DATASET_NAMES = ['PBMC1', 'PBMC2', 'PBMC3','PBMC4']
TOOLS = ['monocle', 'scanpy', 'scvi-tools', 'seurat', 'COTAN']
PARAMS_TUNING = ['default', 'celltypist', 'antibody']def compute_scores(dir, dataset, labels_df, labels_matched, ground_truth_labels):
scores = {}
scores['NMI'] = {}
scores['ARI'] = {}
scores['homogeneity'] = {}
scores['completeness'] = {}
scores['fowlkes_mallows'] = {}
for tool in TOOLS:
scores['NMI'][tool] = normalized_mutual_info_score(labels_pred=labels_df['cluster_'+tool], labels_true=labels_df[f'cluster_{ground_truth_labels}'], average_method='arithmetic')
scores['ARI'][tool] = adjusted_rand_score(labels_pred=labels_df['cluster_'+tool], labels_true=labels_df[f'cluster_{ground_truth_labels}'])
scores['homogeneity'][tool] = homogeneity_score(labels_pred=labels_df['cluster_'+tool], labels_true=labels_df[f'cluster_{ground_truth_labels}'])
scores['completeness'][tool] = completeness_score(labels_pred=labels_df['cluster_'+tool], labels_true=labels_df[f'cluster_{ground_truth_labels}'])
scores['fowlkes_mallows'][tool] = fowlkes_mallows_score(labels_pred=labels_df['cluster_'+tool], labels_true=labels_df[f'cluster_{ground_truth_labels}'])
scores_df = pd.DataFrame(scores)
scores_df.to_csv(f'{dir}{dataset}/scores_{labels_matched}_{ground_truth_labels}.csv')
scores_df.to_latex(f'{dir}{dataset}/scores_{labels_matched}_{ground_truth_labels}.tex')
display(scores_df)
def print_scores(dataset,tuning):
# concat tools labels
labels_df = pd.read_csv(f'{DIR}{dataset}/COTAN/{tuning}/clustering_labels.csv', index_col=0)
labels_df.rename(columns={"cluster": "cluster_COTAN"}, inplace=True)
#print("labels_df size")
#print(labels_df.shape)
for tool in [t for t in TOOLS if t != 'COTAN']:
tool_labels_df = pd.read_csv(f'{DIR}{dataset}/{tool}/{tuning}/clustering_labels.csv', index_col=0)
labels_df = labels_df.merge(tool_labels_df, how='inner', on='cell')
labels_df.rename(columns={"cluster": f"cluster_{tool}"}, inplace=True)
# print("labels_df size"+tool)
# print(labels_df.shape)
# load and concat celltypist labels
celltypist_df = pd.read_csv(f'{DIR}{dataset}/celltypist/celltypist_labels.csv', index_col=0)
celltypist_df.index = celltypist_df.index.str[:-2]
celltypist_df = labels_df.merge(celltypist_df, how='inner', on='cell')
celltypist_df.rename(columns={"cluster.ids": f"cluster_celltypist"}, inplace=True)
celltypist_mapping_df = pd.read_csv(f'{DIR}{dataset}/celltypist/celltypist_mapping.csv', index_col=0)
#print("celltypist_df size")
#print(celltypist_df.shape)
# load and concat protein surface labels
antibody_df = pd.read_csv(f'{DIR}{dataset}/antibody_annotation/antibody_labels_postproc.csv', index_col=0)
antibody_df = labels_df.merge(antibody_df, how='inner', on='cell')
antibody_df.rename(columns={"cluster.ids": f"cluster_antibody"}, inplace=True)
antibody_mapping_df = pd.read_csv(f'{DIR}{dataset}/antibody_annotation/antibody_mapping.csv', index_col=1, encoding='latin1')
#print("antibody_df size")
#print(antibody_df.shape)
# read dataset
adata = sc.read_10x_mtx(
f'{DIR}{dataset}/filtered/10X/',
var_names='gene_symbols',
cache=False
)
# keep only labelled cells
adata.var_names_make_unique()
subset_cells = adata.obs_names.isin(labels_df.index)
adata = adata[subset_cells, :]
mito_genes = adata.var_names.str.startswith('MT-')
# for each cell compute fraction of counts in mito genes vs. all genes
# the `.A1` is only necessary as X is sparse (to transform to a dense array after summing)
adata.obs['percent_mito'] = np.sum(adata[:, mito_genes].X, axis=1).A1 / np.sum(adata.X, axis=1).A1
# add the total counts per cell as observations-annotation to adata
adata.obs['n_counts'] = adata.X.sum(axis=1).A1
sc.pp.normalize_total(adata, target_sum=1e4)
sc.pp.log1p(adata)
sc.pp.highly_variable_genes(adata, min_mean=0.0125, max_mean=3, min_disp=0.5)
adata.raw = adata
adata = adata[:, adata.var.highly_variable]
sc.pp.regress_out(adata, ['n_counts', 'percent_mito'])
sc.pp.scale(adata, max_value=10)
sc.tl.pca(adata, svd_solver='arpack',n_comps=20)
pca_matrix = adata.obsm['X_pca']
scaler = StandardScaler()
scaled_pca_matrix = scaler.fit_transform(pca_matrix)
#Custers number
df = {}
for tool in TOOLS:
df[tool] = labels_df[f'cluster_{tool}'].unique().shape[0]
df_size = pd.DataFrame(df, index=[0])
display(f'{dataset} - number of clusters')
display(df_size)
# compute silhouette score
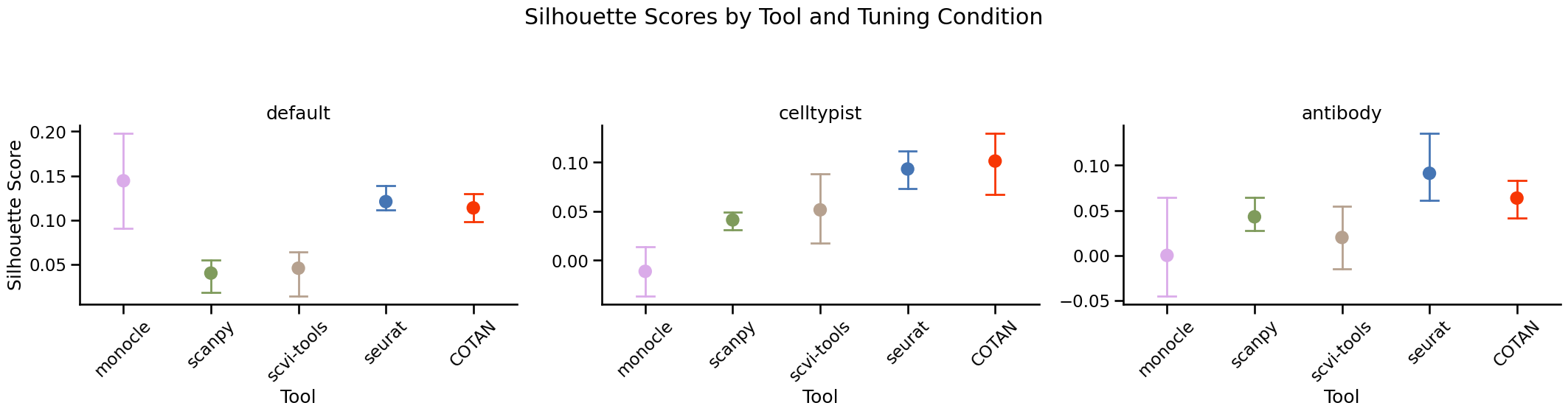
silhouette = {}
for tool in TOOLS:
silhouette[tool] = silhouette_score(scaled_pca_matrix, labels_df[f'cluster_{tool}'])
if tuning=='celltypist':
silhouette['celltypist'] = silhouette_score(scaled_pca_matrix, celltypist_df[f'cluster_celltypist'])
elif tuning=='antibody':
silhouette['antibody'] = silhouette_score(scaled_pca_matrix, antibody_df[f'cluster_antibody'])
silhouette_df = pd.DataFrame(silhouette, index=[0])
silhouette_df.to_csv(f'{DIR}{dataset}/{tuning}_silhouette.csv')
silhouette_df.to_latex(f'{DIR}{dataset}/{tuning}_silhouette.tex')
display(f'{dataset} - Silhuette (higher is better)')
display(silhouette_df)
#From https://evafast.github.io/blog/2019/06/28/example_content/
davies_bouldin = {}
for tool in TOOLS:
davies_bouldin[tool] = davies_bouldin_score(adata.obsm['X_pca'], labels_df[f'cluster_{tool}'])
if tuning=='celltypist':
davies_bouldin['celltypist'] = davies_bouldin_score(adata.obsm['X_pca'], celltypist_df[f'cluster_celltypist'])
elif tuning=='antibody':
davies_bouldin['antibody'] = davies_bouldin_score(adata.obsm['X_pca'], antibody_df[f'cluster_antibody'])
davies_bouldin_df = pd.DataFrame(davies_bouldin, index=[0])
davies_bouldin_df.to_csv(f'{DIR}{dataset}/{tuning}_davies_bouldin.csv')
davies_bouldin_df.to_latex(f'{DIR}{dataset}/{tuning}_davies_bouldin.tex')
display(f'{dataset} - davies_bouldin (lower is better)')
display(davies_bouldin_df)
display(f'{dataset} - matching {tuning} labels' if tuning != 'default' else f'{dataset} - default labels')
# compute scores comparing each tool labels with celltypist labels
if tuning == 'celltypist' or tuning == 'default':
compute_scores(DIR, dataset, celltypist_df, tuning, 'celltypist')
labels = []
labels_titles = []
for tool in TOOLS:
labels.append(celltypist_df[f'cluster_{tool}'].to_list())
labels_titles.append(tool)
labels.append(celltypist_df[f'cluster_celltypist'].map(celltypist_mapping_df['go'].to_dict()).to_list())
labels_titles.append('celltypist')
title = f'{dataset} - matching {tuning} labels' if tuning != 'default' else f'{dataset} - default labels'
sankey_plot(labels=labels, labels_titles=labels_titles, title=title, path=f'{DIR}{dataset}/{tuning}_celltypist.html')
# compute scores comparing each tool labels with protein labels
if tuning == 'antibody' or tuning == 'default':
compute_scores(DIR, dataset, antibody_df, tuning, 'antibody')
labels = []
labels_titles = []
for tool in TOOLS:
labels.append(antibody_df[f'cluster_{tool}'].to_list())
labels_titles.append(tool)
labels.append(antibody_df[f'cluster_antibody'].map(antibody_mapping_df['go'].to_dict()).to_list())
labels_titles.append('antibody')
title = f'{dataset} - matching {tuning} labels' if tuning != 'default' else f'{dataset} - default labels'
sankey_plot(labels=labels, labels_titles=labels_titles, title=title, path=f'{DIR}{dataset}/{tuning}_antibody.html')def print_clustering_data(dataset,tuning):
# concat tools labels
labels_df = pd.read_csv(f'{DIR}{dataset}/COTAN/{tuning}/clustering_labels.csv', index_col=0)
labels_df.rename(columns={"cluster": "cluster_COTAN"}, inplace=True)
display(f'Initial COTAN cluster number:')
display(labels_df.cluster_COTAN.unique().shape[0])
#print("labels_df size")
#print(labels_df.shape)
for tool in [t for t in TOOLS if t != 'COTAN']:
tool_labels_df = pd.read_csv(f'{DIR}{dataset}/{tool}/{tuning}/clustering_labels.csv', index_col=0)
display(f'Initial {tool} cluster number:')
display(labels_df[labels_df.columns[-1]].unique().shape[0])
labels_df = labels_df.merge(tool_labels_df, how='inner', on='cell')
labels_df.rename(columns={"cluster": f"cluster_{tool}"}, inplace=True)
# print("labels_df size"+tool)
# print(labels_df.shape)
if tuning == 'celltypist' or tuning == 'default':
# load and concat celltypist labels
celltypist_df = pd.read_csv(f'{DIR}{dataset}/celltypist/celltypist_labels.csv', index_col=0)
celltypist_df.index = celltypist_df.index.str[:-2]
celltypist_df = labels_df.merge(celltypist_df, how='inner', on='cell')
celltypist_df.rename(columns={"cluster.ids": f"cluster_celltypist"}, inplace=True)
celltypist_mapping_df = pd.read_csv(f'{DIR}{dataset}/celltypist/celltypist_mapping.csv', index_col=0)
#print("celltypist_df size")
#print(celltypist_df.shape)
labels_cluster_celltypist = np.unique(celltypist_df["cluster_celltypist"])
for tool in TOOLS:
labels_cluster_tool = np.unique(celltypist_df[f'cluster_{tool}'])
cm =contingency_matrix(celltypist_df["cluster_celltypist"], celltypist_df[f'cluster_{tool}'])
cm = pd.DataFrame(cm,index=labels_cluster_celltypist,columns=labels_cluster_tool)
display(f'{dataset} - contingency_matrix (rows: cellTypist - cols: {tool})')
display(cm)
if tuning == 'antibody' or tuning == 'default':
#load and concat protein surface labels
antibody_df = pd.read_csv(f'{DIR}{dataset}/antibody_annotation/antibody_labels_postproc.csv', index_col=0)
display("Initial antibody cell/cluster table:")
display(antibody_df["cluster.ids"].value_counts())
antibody_df = labels_df.merge(antibody_df, how='inner', on='cell')
antibody_df.rename(columns={"cluster.ids": f"cluster_antibody"}, inplace=True)
antibody_mapping_df = pd.read_csv(f'{DIR}{dataset}/antibody_annotation/antibody_mapping.csv', index_col=1, encoding='latin1')
labels_cluster_antybody = np.unique(antibody_df["cluster_antibody"])
for tool in TOOLS:
labels_cluster_tool = np.unique(antibody_df[f'cluster_{tool}'])
cm =contingency_matrix(antibody_df["cluster_antibody"], antibody_df[f'cluster_{tool}'])
cm = pd.DataFrame(cm,index=labels_cluster_antybody,columns=labels_cluster_tool)
display(f'{dataset} - contingency_matrix (rows: antibody - cols: {tool})')
display(cm)
Data summary information
Default parameters
print_clustering_data(tuning = 'default',dataset="PBMC1")'Initial COTAN cluster number:'14'Initial monocle cluster number:'14'Initial scanpy cluster number:'3'Initial scvi-tools cluster number:'18'Initial seurat cluster number:'13'PBMC1 - contingency_matrix (rows: cellTypist - cols: monocle)'| 1 | 2 | 3 | |
|---|---|---|---|
| 1 | 8 | 970 | 1 |
| 2 | 943 | 0 | 0 |
| 3 | 47 | 0 | 0 |
| 4 | 0 | 78 | 0 |
| 5 | 309 | 0 | 0 |
| 6 | 0 | 0 | 142 |
| 7 | 82 | 0 | 0 |
| 8 | 278 | 0 | 1 |
| 9 | 81 | 0 | 0 |
| 10 | 0 | 171 | 0 |
| 11 | 70 | 0 | 0 |
| 12 | 240 | 0 | 0 |
| 13 | 0 | 28 | 0 |
| 14 | 0 | 0 | 155 |
| 15 | 2 | 4 | 0 |
'PBMC1 - contingency_matrix (rows: cellTypist - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 246 | 0 | 267 | 0 | 263 | 0 | 200 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| 2 | 88 | 321 | 0 | 281 | 1 | 0 | 0 | 241 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 8 | 0 | 0 |
| 3 | 45 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 76 | 0 |
| 5 | 250 | 5 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 52 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 138 | 0 | 0 | 0 | 0 | 0 |
| 7 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19 | 0 | 37 | 0 | 18 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 | 0 | 0 | 263 | 0 | 0 | 0 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 2 | 2 | 0 | 5 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 66 | 0 | 0 |
| 10 | 0 | 0 | 75 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 91 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 65 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 82 | 0 | 59 | 0 | 86 | 0 | 3 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 154 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: cellTypist - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 659 | 0 | 0 | 0 | 289 | 0 | 1 | 0 | 0 | 30 | 0 | 0 | 0 |
| 2 | 0 | 485 | 48 | 402 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 6 | 0 |
| 3 | 0 | 1 | 41 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 72 | 0 | 0 | 0 |
| 5 | 0 | 5 | 288 | 12 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 139 | 0 | 0 | 0 | 0 |
| 7 | 0 | 0 | 58 | 0 | 0 | 23 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 | 279 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 73 | 0 |
| 10 | 1 | 0 | 0 | 0 | 78 | 0 | 0 | 0 | 0 | 0 | 92 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 67 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 1 | 48 | 1 | 0 | 189 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 |
| 14 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 147 | 7 | 0 | 0 | 0 | 0 |
| 15 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: cellTypist - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 616 | 361 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| 2 | 798 | 145 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 1 | 46 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 74 | 0 |
| 5 | 0 | 309 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 142 | 0 | 0 | 0 |
| 7 | 0 | 55 | 0 | 0 | 0 | 27 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 | 0 | 274 | 5 | 0 | 0 | 0 | 0 | 0 |
| 9 | 78 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 43 | 0 | 0 | 0 | 0 | 128 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 5 | 65 | 0 | 0 | 0 | 0 | 0 |
| 12 | 7 | 69 | 0 | 0 | 0 | 164 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 27 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 153 | 2 | 0 | 0 | 0 |
| 15 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: cellTypist - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 35 | 0 | 11 | 648 | 284 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 807 | 129 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 37 | 2 | 8 |
| 4 | 0 | 73 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 308 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 142 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 1 | 0 | 0 | 56 |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 55 | 4 | 220 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 34 | 1 | 0 | 0 | 45 | 1 |
| 10 | 0 | 1 | 146 | 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 67 | 1 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 175 | 0 | 1 | 5 | 56 |
| 13 | 0 | 1 | 0 | 0 | 0 | 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 152 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
'Initial antibody cell/cluster table:'cluster.ids
7 1338
8 876
3 748
4 341
9 331
5 211
1 202
6 131
2 62
10 51
12 16
Name: count, dtype: int64'PBMC1 - contingency_matrix (rows: antibody - cols: monocle)'| 1 | 2 | 3 | |
|---|---|---|---|
| 1 | 161 | 0 | 0 |
| 2 | 0 | 43 | 3 |
| 3 | 600 | 0 | 0 |
| 4 | 262 | 1 | 0 |
| 5 | 158 | 0 | 0 |
| 6 | 1 | 86 | 0 |
| 7 | 10 | 1115 | 0 |
| 8 | 812 | 1 | 1 |
| 9 | 1 | 0 | 294 |
| 10 | 44 | 0 | 0 |
| 12 | 10 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7 | 6 | 0 | 4 | 0 | 0 | 0 | 9 | 0 | 23 | 0 | 7 | 0 | 32 | 0 | 73 | 0 | 0 |
| 2 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 3 | 0 | 38 | 0 | 0 | 0 |
| 3 | 366 | 33 | 0 | 5 | 0 | 0 | 0 | 28 | 0 | 43 | 0 | 122 | 0 | 3 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 | 1 | 249 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 64 | 0 | 15 | 0 | 68 | 0 | 0 | 0 | 0 |
| 6 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 56 | 26 |
| 7 | 1 | 0 | 319 | 0 | 267 | 0 | 267 | 1 | 198 | 0 | 0 | 0 | 0 | 0 | 52 | 0 | 19 | 1 |
| 8 | 18 | 288 | 0 | 277 | 0 | 1 | 0 | 214 | 0 | 3 | 1 | 5 | 0 | 2 | 0 | 4 | 0 | 1 |
| 9 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 156 | 0 | 137 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 36 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 6 | 12 | 5 | 0 | 62 | 0 | 0 | 0 | 0 | 0 | 76 | 0 |
| 2 | 2 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 1 | 0 | 39 | 0 | 0 |
| 3 | 0 | 49 | 441 | 66 | 0 | 44 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 1 | 0 | 0 | 0 | 0 | 0 | 262 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 0 | 2 | 12 | 1 | 0 | 141 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| 6 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 56 | 1 | 0 | 26 |
| 7 | 655 | 3 | 0 | 0 | 368 | 0 | 1 | 0 | 0 | 45 | 52 | 0 | 1 |
| 8 | 0 | 438 | 17 | 349 | 0 | 4 | 3 | 0 | 0 | 0 | 0 | 2 | 1 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 148 | 144 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 1 | 0 | 0 | 37 | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 93 | 17 | 0 | 0 | 0 | 51 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 1 | 4 | 0 | 0 | 1 | 1 | 39 | 0 | 0 |
| 3 | 23 | 540 | 0 | 0 | 0 | 37 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 1 | 0 | 260 | 2 | 0 | 0 | 0 | 0 | 0 |
| 5 | 1 | 28 | 0 | 0 | 0 | 129 | 0 | 0 | 0 | 0 | 0 |
| 6 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 56 | 25 |
| 7 | 2 | 1 | 611 | 402 | 1 | 0 | 0 | 0 | 89 | 18 | 1 |
| 8 | 766 | 41 | 0 | 0 | 1 | 4 | 1 | 0 | 0 | 0 | 1 |
| 9 | 0 | 0 | 0 | 0 | 2 | 0 | 151 | 142 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 8 | 36 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 8 | 2 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 36 | 54 | 0 | 1 | 59 | 11 |
| 2 | 1 | 1 | 40 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 39 | 0 | 38 | 33 | 490 |
| 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 1 | 213 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 135 | 0 | 0 | 0 | 22 |
| 6 | 0 | 56 | 1 | 1 | 1 | 25 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| 7 | 33 | 17 | 118 | 668 | 284 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 1 |
| 8 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 9 | 4 | 0 | 0 | 765 | 34 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 150 | 143 | 0 | 0 | 2 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 38 | 2 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 1 | 5 | 0 | 0 | 0 |
print_clustering_data(tuning = 'default',dataset="PBMC2")'Initial COTAN cluster number:'17'Initial monocle cluster number:'17'Initial scanpy cluster number:'2'Initial scvi-tools cluster number:'18'Initial seurat cluster number:'20'PBMC2 - contingency_matrix (rows: cellTypist - cols: monocle)'| 1 | 2 | |
|---|---|---|
| 1 | 230 | 1 |
| 2 | 427 | 0 |
| 3 | 2139 | 3 |
| 4 | 700 | 7 |
| 5 | 316 | 0 |
| 6 | 0 | 93 |
| 7 | 0 | 567 |
| 8 | 674 | 0 |
| 9 | 0 | 186 |
| 10 | 52 | 0 |
| 11 | 0 | 228 |
| 12 | 0 | 204 |
| 13 | 0 | 48 |
| 14 | 0 | 14 |
| 15 | 80 | 0 |
| 16 | 0 | 6 |
'PBMC2 - contingency_matrix (rows: cellTypist - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 22 | 1 | 2 | 5 | 50 | 0 | 148 | 0 | 0 | 0 | 0 |
| 2 | 0 | 91 | 0 | 273 | 0 | 0 | 0 | 3 | 2 | 0 | 56 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| 3 | 942 | 508 | 21 | 183 | 0 | 0 | 0 | 21 | 295 | 2 | 100 | 42 | 0 | 26 | 1 | 0 | 0 | 1 |
| 4 | 0 | 0 | 0 | 1 | 0 | 463 | 8 | 2 | 0 | 230 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 266 | 0 | 0 | 42 | 0 | 0 | 6 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 88 | 0 | 0 |
| 7 | 0 | 0 | 0 | 0 | 466 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 86 | 0 | 0 | 0 |
| 8 | 2 | 1 | 558 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 110 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 174 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 50 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 228 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 204 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 |
| 15 | 0 | 25 | 0 | 46 | 0 | 0 | 0 | 1 | 2 | 1 | 2 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC2 - contingency_matrix (rows: cellTypist - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 222 | 1 | 2 | 0 | 0 | 4 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 331 | 4 | 1 | 0 | 0 | 0 | 7 | 10 | 53 | 0 | 0 | 1 | 0 | 10 | 4 | 0 | 6 | 0 | 0 | 0 |
| 3 | 391 | 733 | 2 | 1 | 19 | 1 | 41 | 304 | 158 | 185 | 0 | 2 | 43 | 90 | 75 | 0 | 71 | 0 | 0 | 26 |
| 4 | 1 | 0 | 675 | 1 | 0 | 8 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19 | 0 | 0 |
| 5 | 7 | 0 | 1 | 0 | 0 | 0 | 11 | 9 | 66 | 0 | 0 | 151 | 71 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 78 | 0 | 0 | 3 | 0 |
| 7 | 0 | 0 | 0 | 564 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 2 | 0 | 1 | 561 | 0 | 103 | 1 | 0 | 2 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 175 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 2 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 44 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 228 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 204 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 58 | 0 | 1 | 0 | 0 | 0 | 5 | 5 | 1 | 5 | 0 | 0 | 1 | 3 | 0 | 0 | 1 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC2 - contingency_matrix (rows: cellTypist - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 4 | 6 | 0 | 0 | 0 | 219 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| 2 | 0 | 400 | 23 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| 3 | 1151 | 352 | 550 | 0 | 2 | 15 | 69 | 1 | 0 | 0 | 0 | 2 | 0 | 0 |
| 4 | 0 | 0 | 2 | 635 | 0 | 0 | 3 | 7 | 0 | 0 | 1 | 59 | 0 | 0 |
| 5 | 0 | 42 | 0 | 0 | 0 | 0 | 7 | 0 | 267 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 82 | 0 |
| 7 | 0 | 0 | 0 | 0 | 567 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 8 | 4 | 2 | 0 | 0 | 541 | 119 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 172 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 49 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 83 | 0 | 0 | 145 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 201 | 1 | 0 | 2 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 |
| 15 | 0 | 0 | 77 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 |
'PBMC2 - contingency_matrix (rows: cellTypist - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 3 | 4 | 220 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 416 | 9 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 2 | 1186 | 847 | 31 | 72 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 4 | 639 | 56 | 0 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 5 | 1 |
| 5 | 0 | 0 | 0 | 39 | 0 | 7 | 270 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 86 | 0 | 7 | 0 | 0 | 0 | 0 | 0 |
| 7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 88 | 300 | 174 | 0 | 0 | 3 | 1 |
| 8 | 0 | 0 | 568 | 9 | 1 | 96 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 154 | 24 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 |
| 10 | 1 | 51 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 145 | 69 | 14 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 22 | 180 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 1 |
| 15 | 0 | 1 | 0 | 6 | 72 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 0 |
'Initial antibody cell/cluster table:'cluster.ids
4 1510
11 1130
8 695
12 570
6 424
13 275
5 197
2 150
10 122
3 84
7 76
Name: count, dtype: int64'PBMC2 - contingency_matrix (rows: antibody - cols: monocle)'| 1 | 2 | |
|---|---|---|
| 2 | 0 | 147 |
| 3 | 60 | 20 |
| 4 | 1480 | 13 |
| 5 | 196 | 0 |
| 6 | 416 | 1 |
| 7 | 68 | 7 |
| 8 | 681 | 5 |
| 10 | 0 | 119 |
| 11 | 1115 | 9 |
| 12 | 566 | 4 |
| 13 | 0 | 271 |
'PBMC2 - contingency_matrix (rows: antibody - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 141 | 0 | 2 | 0 | 0 | 0 |
| 3 | 0 | 1 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 54 | 0 | 0 | 18 | 2 | 1 | 0 | 0 | 0 |
| 4 | 89 | 588 | 0 | 478 | 0 | 0 | 0 | 56 | 63 | 0 | 194 | 10 | 1 | 3 | 3 | 4 | 3 | 1 |
| 5 | 7 | 1 | 14 | 2 | 0 | 0 | 0 | 118 | 2 | 2 | 4 | 23 | 0 | 23 | 0 | 0 | 0 | 0 |
| 6 | 0 | 2 | 9 | 13 | 0 | 0 | 1 | 128 | 1 | 0 | 3 | 124 | 0 | 136 | 0 | 0 | 0 | 0 |
| 7 | 1 | 3 | 27 | 3 | 0 | 0 | 0 | 6 | 1 | 0 | 1 | 23 | 0 | 3 | 0 | 5 | 2 | 0 |
| 8 | 0 | 0 | 0 | 0 | 0 | 459 | 4 | 0 | 0 | 220 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 |
| 10 | 0 | 0 | 0 | 0 | 107 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 3 | 6 | 0 | 0 |
| 11 | 843 | 26 | 5 | 5 | 0 | 0 | 5 | 4 | 229 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 1 | 0 |
| 12 | 2 | 0 | 522 | 0 | 0 | 0 | 2 | 1 | 1 | 2 | 0 | 21 | 0 | 17 | 1 | 0 | 1 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 263 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 1 | 0 |
'PBMC2 - contingency_matrix (rows: antibody - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 142 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 0 | 55 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 |
| 4 | 758 | 143 | 0 | 4 | 0 | 0 | 30 | 125 | 259 | 52 | 0 | 5 | 4 | 54 | 26 | 4 | 23 | 0 | 3 | 3 |
| 5 | 4 | 8 | 1 | 0 | 5 | 0 | 27 | 4 | 6 | 0 | 0 | 36 | 105 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 4 | 0 | 0 | 0 | 17 | 0 | 270 | 2 | 9 | 0 | 0 | 112 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 1 | 0 | 1 | 31 | 0 | 27 | 2 | 1 | 1 | 0 | 5 | 0 | 0 | 0 | 4 | 0 | 0 | 2 | 0 |
| 8 | 0 | 0 | 622 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 56 | 1 | 0 |
| 10 | 0 | 0 | 0 | 111 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| 11 | 14 | 581 | 0 | 1 | 5 | 5 | 2 | 191 | 5 | 138 | 0 | 0 | 1 | 50 | 51 | 2 | 54 | 0 | 1 | 23 |
| 12 | 0 | 2 | 1 | 1 | 520 | 2 | 34 | 1 | 0 | 1 | 0 | 0 | 6 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| 13 | 0 | 0 | 0 | 7 | 0 | 263 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
'PBMC2 - contingency_matrix (rows: antibody - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 139 | 0 | 0 | 0 | 0 |
| 3 | 0 | 0 | 1 | 9 | 2 | 1 | 1 | 0 | 0 | 18 | 0 | 48 | 0 | 0 |
| 4 | 65 | 756 | 607 | 0 | 4 | 0 | 10 | 0 | 42 | 0 | 0 | 2 | 4 | 3 |
| 5 | 6 | 6 | 2 | 0 | 0 | 12 | 65 | 0 | 104 | 0 | 0 | 1 | 0 | 0 |
| 6 | 0 | 22 | 3 | 0 | 0 | 7 | 267 | 0 | 118 | 0 | 0 | 0 | 0 | 0 |
| 7 | 2 | 7 | 5 | 0 | 0 | 21 | 30 | 0 | 3 | 0 | 0 | 0 | 5 | 2 |
| 8 | 0 | 0 | 0 | 627 | 0 | 0 | 0 | 3 | 0 | 2 | 1 | 52 | 0 | 1 |
| 10 | 0 | 0 | 0 | 0 | 113 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 3 | 0 |
| 11 | 1073 | 4 | 35 | 0 | 1 | 1 | 1 | 4 | 1 | 0 | 1 | 0 | 2 | 1 |
| 12 | 9 | 0 | 0 | 0 | 1 | 512 | 44 | 1 | 0 | 0 | 1 | 1 | 0 | 1 |
| 13 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 159 | 1 | 0 | 103 | 0 | 0 | 1 |
'PBMC2 - contingency_matrix (rows: antibody - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 137 | 4 | 0 | 2 | 3 | 1 | 0 | 0 | 0 | 0 |
| 3 | 8 | 49 | 1 | 2 | 0 | 1 | 0 | 1 | 17 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 76 | 1235 | 110 | 16 | 44 | 0 | 0 | 4 | 3 | 0 | 1 | 3 | 0 | 1 | 0 |
| 5 | 0 | 0 | 19 | 6 | 1 | 65 | 105 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 11 | 23 | 1 | 261 | 120 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 0 | 32 | 16 | 1 | 15 | 4 | 0 | 0 | 5 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| 8 | 630 | 50 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 0 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 5 | 1 | 76 | 33 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 1082 | 27 | 4 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 1 | 3 | 1 |
| 12 | 0 | 1 | 526 | 0 | 0 | 38 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 5 | 0 | 0 | 1 | 102 | 29 | 133 |
print_clustering_data(tuning = 'default',dataset="PBMC3")'Initial COTAN cluster number:'32'Initial monocle cluster number:'32'Initial scanpy cluster number:'3'Initial scvi-tools cluster number:'22'Initial seurat cluster number:'17'PBMC3 - contingency_matrix (rows: cellTypist - cols: monocle)'| 1 | 2 | 3 | |
|---|---|---|---|
| 1 | 3021 | 0 | 0 |
| 2 | 1 | 1471 | 0 |
| 3 | 6 | 1 | 655 |
| 4 | 1100 | 0 | 0 |
| 5 | 1183 | 26 | 33 |
| 6 | 0 | 156 | 0 |
| 7 | 1112 | 1 | 0 |
| 8 | 484 | 0 | 0 |
| 9 | 0 | 0 | 396 |
| 10 | 0 | 408 | 0 |
| 11 | 430 | 0 | 0 |
| 12 | 0 | 0 | 8 |
| 13 | 233 | 0 | 0 |
| 14 | 111 | 0 | 0 |
| 15 | 4 | 16 | 0 |
| 16 | 0 | 11 | 0 |
| 17 | 0 | 8 | 0 |
| 18 | 0 | 57 | 0 |
| 19 | 0 | 12 | 0 |
'PBMC3 - contingency_matrix (rows: cellTypist - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ... | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1401 | 0 | 153 | 0 | 0 | 29 | 26 | 534 | 121 | 429 | ... | 1 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 28 | 0 |
| 2 | 0 | 0 | 0 | 0 | 816 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 2 | 0 | 227 | 0 | 5 | 0 | 35 | 0 | 0 |
| 3 | 0 | 0 | 0 | 543 | 0 | 0 | 0 | 0 | 5 | 0 | ... | 0 | 0 | 0 | 0 | 111 | 0 | 0 | 2 | 0 | 0 |
| 4 | 26 | 0 | 0 | 0 | 0 | 806 | 12 | 5 | 29 | 6 | ... | 0 | 0 | 155 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 5 | 0 | 961 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | ... | 216 | 0 | 0 | 29 | 33 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 147 | 0 | 0 | 0 | 0 |
| 7 | 0 | 0 | 683 | 0 | 0 | 0 | 7 | 128 | 236 | 15 | ... | 0 | 0 | 6 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 | 423 | 0 | 32 | 0 | ... | 4 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 395 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 90 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 311 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| 11 | 0 | 2 | 0 | 0 | 0 | 0 | 281 | 2 | 54 | 0 | ... | 4 | 0 | 42 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 |
| 13 | 12 | 0 | 116 | 0 | 0 | 0 | 1 | 52 | 9 | 11 | ... | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 5 | 1 | 0 | 0 | 0 | 5 | 0 | 3 | 0 | ... | 97 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 20 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 57 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 |
19 rows × 22 columns
'PBMC3 - contingency_matrix (rows: cellTypist - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 740 | 1603 | 2 | 1 | 1 | 40 | 86 | 23 | 248 | 1 | 197 | 79 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 1460 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 8 | 0 | 0 | 0 | 0 |
| 3 | 0 | 0 | 4 | 2 | 655 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 6 | 11 | 0 | 1 | 1 | 912 | 162 | 2 | 1 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 0 | 0 | 29 | 1165 | 31 | 0 | 7 | 1 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 1 | 0 |
| 6 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 146 | 0 | 0 | 0 | 0 |
| 7 | 883 | 14 | 1 | 0 | 0 | 1 | 25 | 29 | 80 | 0 | 70 | 10 | 0 | 0 | 0 | 0 | 0 |
| 8 | 24 | 0 | 0 | 6 | 0 | 1 | 5 | 313 | 0 | 0 | 0 | 135 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 396 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 97 | 0 | 0 | 0 | 0 | 0 | 0 | 311 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 10 | 0 | 0 | 2 | 0 | 2 | 408 | 2 | 0 | 0 | 1 | 4 | 0 | 1 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 174 | 26 | 0 | 0 | 0 | 0 | 3 | 0 | 25 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 1 | 19 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 90 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 20 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 55 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC3 - contingency_matrix (rows: cellTypist - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1740 | 415 | 0 | 0 | 681 | 20 | 162 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 1013 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 227 | 0 | 229 | 3 | 0 | 0 | 0 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 535 | 0 | 2 | 0 | 1 | 2 | 0 | 0 | 120 | 0 | 0 |
| 4 | 16 | 6 | 0 | 0 | 3 | 886 | 188 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 5 | 0 | 1 | 4 | 1043 | 0 | 0 | 2 | 1 | 1 | 0 | 0 | 24 | 136 | 0 | 0 | 30 | 0 | 0 |
| 6 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 149 | 0 | 0 | 0 |
| 7 | 0 | 980 | 0 | 0 | 108 | 0 | 18 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 18 | 0 | 3 | 0 | 0 | 8 | 0 | 454 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 57 | 0 | 336 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| 10 | 0 | 0 | 84 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 319 | 1 | 0 | 4 | 0 | 0 | 0 | 0 |
| 11 | 0 | 14 | 0 | 0 | 0 | 2 | 411 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 13 | 13 | 5 | 0 | 0 | 208 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 109 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 20 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 52 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 |
'PBMC3 - contingency_matrix (rows: cellTypist - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ... | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 48 | 6 | 205 | 1790 | 15 | 4 | 53 | 199 | 154 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | ... | 67 | 125 | 220 | 131 | 0 | 0 | 2 | 0 | 0 | 0 |
| 4 | 0 | 152 | 1 | 89 | 18 | 805 | 0 | 2 | 7 | 20 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | ... | 3 | 0 | 3 | 0 | 30 | 4 | 138 | 1025 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 12 | 9 | 26 | 0 | 0 | 4 | 431 | 546 | 27 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 8 | 0 | 4 | 465 | 0 | 0 | 0 | 0 | 1 | 8 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 30 | 15 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 83 | 272 | 2 | 9 | 0 | 1 | 0 | 0 | 7 | 8 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 1 | 32 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 |
| 13 | 0 | 0 | 0 | 4 | 5 | 0 | 139 | 3 | 1 | 6 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 86 | 17 | 6 | 0 | 0 |
| 15 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 52 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 11 | 0 |
19 rows × 32 columns
'Initial antibody cell/cluster table:'cluster.ids
9 2220
10 1635
7 1271
13 1067
5 1010
12 909
6 744
2 271
4 214
14 168
3 149
23 133
22 71
Name: count, dtype: int64'PBMC3 - contingency_matrix (rows: antibody - cols: monocle)'| 1 | 2 | 3 | |
|---|---|---|---|
| 2 | 3 | 265 | 0 |
| 3 | 132 | 13 | 3 |
| 4 | 209 | 2 | 1 |
| 5 | 993 | 14 | 1 |
| 6 | 741 | 0 | 0 |
| 7 | 4 | 1226 | 0 |
| 9 | 2201 | 9 | 1 |
| 10 | 1616 | 12 | 3 |
| 12 | 905 | 2 | 1 |
| 13 | 2 | 43 | 1002 |
| 14 | 141 | 23 | 1 |
| 22 | 70 | 1 | 0 |
| 23 | 130 | 0 | 0 |
'PBMC3 - contingency_matrix (rows: antibody - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ... | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 0 | 1 | 0 | 0 | 17 | 0 | 0 | 0 | 0 | 0 | ... | 1 | 223 | 0 | 25 | 0 | 0 | 1 | 0 | 0 | 0 |
| 3 | 0 | 14 | 2 | 0 | 0 | 1 | 5 | 0 | 3 | 0 | ... | 106 | 12 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| 4 | 4 | 0 | 5 | 0 | 0 | 50 | 25 | 10 | 28 | 15 | ... | 1 | 0 | 57 | 0 | 1 | 2 | 0 | 0 | 1 | 0 |
| 5 | 0 | 912 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | ... | 80 | 10 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 1 |
| 6 | 1 | 0 | 8 | 0 | 0 | 9 | 433 | 11 | 75 | 1 | ... | 3 | 0 | 151 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 0 | 1 | 0 | 735 | 0 | 0 | 1 | 0 | 0 | ... | 0 | 20 | 0 | 167 | 0 | 28 | 2 | 0 | 0 | 1 |
| 9 | 92 | 0 | 808 | 1 | 0 | 1 | 26 | 593 | 323 | 196 | ... | 0 | 3 | 19 | 1 | 0 | 5 | 0 | 0 | 7 | 0 |
| 10 | 1252 | 2 | 4 | 0 | 0 | 15 | 3 | 18 | 4 | 205 | ... | 3 | 1 | 5 | 3 | 6 | 4 | 2 | 0 | 20 | 2 |
| 12 | 25 | 1 | 0 | 0 | 1 | 737 | 34 | 0 | 8 | 2 | ... | 6 | 0 | 40 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 873 | 1 | 0 | 1 | 0 | 1 | 0 | ... | 0 | 6 | 0 | 1 | 128 | 2 | 2 | 30 | 0 | 0 |
| 14 | 0 | 17 | 0 | 0 | 0 | 0 | 4 | 0 | 2 | 1 | ... | 116 | 20 | 0 | 1 | 1 | 0 | 3 | 0 | 0 | 0 |
| 22 | 10 | 0 | 9 | 0 | 0 | 0 | 2 | 18 | 6 | 19 | ... | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| 23 | 0 | 0 | 90 | 0 | 0 | 0 | 0 | 18 | 1 | 2 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
13 rows × 22 columns
'PBMC3 - contingency_matrix (rows: antibody - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 0 | 0 | 42 | 2 | 0 | 0 | 0 | 0 | 0 | 223 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 3 | 3 | 0 | 1 | 48 | 2 | 1 | 6 | 1 | 0 | 12 | 0 | 0 | 0 | 74 | 0 | 0 | 0 |
| 4 | 18 | 8 | 1 | 0 | 2 | 76 | 80 | 18 | 0 | 0 | 7 | 0 | 2 | 0 | 0 | 0 | 0 |
| 5 | 0 | 0 | 1 | 979 | 1 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 14 | 0 | 2 | 1 |
| 6 | 14 | 1 | 1 | 1 | 0 | 23 | 464 | 222 | 0 | 0 | 6 | 6 | 0 | 3 | 0 | 0 | 0 |
| 7 | 2 | 0 | 1182 | 0 | 0 | 0 | 0 | 0 | 0 | 18 | 0 | 0 | 26 | 0 | 1 | 1 | 0 |
| 9 | 1524 | 275 | 1 | 0 | 2 | 1 | 26 | 47 | 171 | 3 | 134 | 22 | 5 | 0 | 0 | 0 | 0 |
| 10 | 52 | 1281 | 1 | 6 | 4 | 15 | 4 | 2 | 149 | 2 | 101 | 4 | 6 | 0 | 2 | 2 | 0 |
| 12 | 0 | 5 | 2 | 4 | 1 | 816 | 67 | 5 | 2 | 0 | 1 | 5 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 31 | 1 | 995 | 0 | 1 | 1 | 8 | 7 | 0 | 0 | 2 | 0 | 1 | 0 | 0 |
| 14 | 0 | 0 | 1 | 129 | 1 | 0 | 0 | 3 | 0 | 20 | 0 | 1 | 0 | 7 | 3 | 0 | 0 |
| 22 | 39 | 14 | 0 | 0 | 0 | 0 | 0 | 4 | 4 | 0 | 6 | 3 | 0 | 0 | 1 | 0 | 0 |
| 23 | 111 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC3 - contingency_matrix (rows: antibody - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 0 | 0 | 18 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 226 | 20 | 1 | 0 | 0 | 0 | 1 | 0 |
| 3 | 0 | 3 | 0 | 25 | 0 | 1 | 1 | 0 | 2 | 0 | 12 | 0 | 101 | 0 | 0 | 2 | 0 | 1 |
| 4 | 11 | 28 | 0 | 0 | 12 | 48 | 97 | 0 | 12 | 0 | 0 | 0 | 1 | 0 | 2 | 1 | 0 | 0 |
| 5 | 0 | 0 | 0 | 980 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 1 | 14 | 0 | 0 | 1 | 0 | 2 |
| 6 | 1 | 22 | 0 | 0 | 6 | 16 | 468 | 0 | 225 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 2 | 873 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 22 | 139 | 0 | 164 | 27 | 0 | 2 | 1 |
| 9 | 129 | 1302 | 0 | 0 | 716 | 0 | 31 | 1 | 23 | 0 | 3 | 1 | 0 | 0 | 5 | 0 | 0 | 0 |
| 10 | 1543 | 8 | 1 | 2 | 46 | 3 | 6 | 0 | 2 | 0 | 0 | 3 | 3 | 0 | 4 | 4 | 2 | 4 |
| 12 | 6 | 0 | 1 | 0 | 3 | 816 | 72 | 0 | 4 | 0 | 0 | 1 | 4 | 0 | 0 | 1 | 0 | 0 |
| 13 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 549 | 1 | 327 | 7 | 30 | 0 | 1 | 2 | 126 | 2 | 0 |
| 14 | 0 | 1 | 0 | 14 | 1 | 0 | 0 | 0 | 3 | 0 | 20 | 1 | 121 | 0 | 0 | 1 | 3 | 0 |
| 22 | 12 | 19 | 0 | 0 | 34 | 0 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 23 | 0 | 4 | 0 | 0 | 126 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC3 - contingency_matrix (rows: antibody - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ... | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 14 | 0 | 1 | 2 | 0 | 1 |
| 3 | 1 | 0 | 1 | 3 | 0 | 1 | 0 | 0 | 1 | 0 | ... | 1 | 0 | 0 | 0 | 0 | 72 | 28 | 25 | 1 | 0 |
| 4 | 3 | 29 | 15 | 13 | 15 | 50 | 2 | 9 | 15 | 44 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 |
| 5 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 1 | 0 | 12 | 11 | 2 | 968 | 2 | 0 |
| 6 | 62 | 357 | 227 | 8 | 1 | 13 | 1 | 14 | 11 | 4 | ... | 0 | 0 | 0 | 0 | 0 | 1 | 15 | 0 | 18 | 0 |
| 7 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 2 |
| 9 | 1 | 21 | 28 | 159 | 158 | 0 | 30 | 440 | 716 | 134 | ... | 0 | 0 | 1 | 0 | 0 | 0 | 8 | 0 | 2 | 0 |
| 10 | 0 | 5 | 3 | 3 | 1550 | 3 | 0 | 6 | 2 | 5 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 2 | 2 | 2 |
| 12 | 6 | 47 | 4 | 79 | 5 | 739 | 0 | 0 | 1 | 3 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 6 | 0 |
| 13 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | ... | 65 | 116 | 233 | 128 | 1 | 1 | 0 | 0 | 8 | 2 |
| 14 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 4 | 116 | 14 | 0 | 3 |
| 22 | 0 | 0 | 2 | 2 | 19 | 0 | 1 | 8 | 6 | 9 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 23 | 0 | 0 | 0 | 4 | 0 | 0 | 109 | 4 | 0 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
13 rows × 32 columns
print_clustering_data(tuning = 'default',dataset="PBMC4")'Initial COTAN cluster number:'24'Initial monocle cluster number:'24'Initial scanpy cluster number:'3'Initial scvi-tools cluster number:'22'Initial seurat cluster number:'16'PBMC4 - contingency_matrix (rows: cellTypist - cols: monocle)'| 1 | 2 | 3 | |
|---|---|---|---|
| 1 | 407 | 0 | 0 |
| 2 | 11 | 0 | 797 |
| 3 | 1330 | 1 | 0 |
| 4 | 108 | 0 | 0 |
| 5 | 9 | 2178 | 13 |
| 6 | 308 | 0 | 0 |
| 7 | 77 | 0 | 0 |
| 8 | 538 | 0 | 0 |
| 9 | 358 | 1 | 0 |
| 10 | 0 | 307 | 0 |
| 11 | 1 | 1 | 222 |
| 12 | 0 | 28 | 0 |
| 13 | 8 | 3 | 2 |
| 14 | 106 | 0 | 0 |
| 15 | 0 | 92 | 1 |
| 16 | 0 | 59 | 0 |
'PBMC4 - contingency_matrix (rows: cellTypist - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ... | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 384 | 1 | 0 | ... | 0 | 6 | 0 | 0 | 0 | 1 | 5 | 0 | 0 | 0 |
| 2 | 0 | 673 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 34 |
| 3 | 300 | 0 | 0 | 0 | 496 | 385 | 0 | 1 | 0 | 0 | ... | 136 | 10 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 10 | 0 | ... | 0 | 7 | 0 | 0 | 0 | 0 | 85 | 0 | 0 | 0 |
| 5 | 1 | 0 | 596 | 456 | 0 | 0 | 427 | 0 | 0 | 281 | ... | 0 | 0 | 0 | 169 | 76 | 145 | 0 | 48 | 0 | 0 |
| 6 | 7 | 0 | 0 | 0 | 4 | 3 | 0 | 0 | 0 | 0 | ... | 8 | 99 | 187 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | ... | 0 | 67 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 |
| 8 | 462 | 0 | 0 | 0 | 8 | 46 | 0 | 1 | 0 | 0 | ... | 19 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 348 | 0 | ... | 1 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 74 | 0 | 0 | 1 | 0 | 0 | 2 | ... | 0 | 0 | 0 | 10 | 2 | 1 | 0 | 1 | 0 | 0 |
| 11 | 0 | 46 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 5 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 0 | 0 |
| 13 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 42 | 0 | 0 | 0 | 1 | 13 | 0 | 1 | 0 | 0 | ... | 47 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | ... | 0 | 0 | 0 | 2 | 88 | 2 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 59 | 0 |
16 rows × 22 columns
'PBMC4 - contingency_matrix (rows: cellTypist - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 4 | 1 | 0 | 0 | 1 | 0 | 315 | 0 | 0 | 0 | 86 | 0 | 0 | 0 |
| 2 | 800 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| 3 | 0 | 939 | 357 | 2 | 0 | 0 | 20 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 12 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 105 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 5 | 4 | 0 | 1 | 805 | 746 | 413 | 0 | 4 | 2 | 1 | 167 | 15 | 1 | 41 | 0 | 0 |
| 6 | 0 | 8 | 8 | 0 | 0 | 0 | 291 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 0 | 3 | 0 | 0 | 0 | 67 | 1 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 31 | 483 | 0 | 0 | 0 | 3 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 15 |
| 9 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 355 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 14 | 0 | 94 | 1 | 0 | 0 | 194 | 4 | 0 | 0 | 0 | 0 | 0 |
| 11 | 217 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 0 | 0 |
| 13 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 |
| 14 | 1 | 9 | 89 | 0 | 0 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| 15 | 0 | 0 | 0 | 2 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 87 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 58 | 0 |
'PBMC4 - contingency_matrix (rows: cellTypist - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 0 | 0 | 0 | 0 | 0 | 398 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 632 | 0 | 1 | 0 | 122 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 52 | 0 |
| 3 | 356 | 854 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 116 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 107 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 2 | 0 | 780 | 723 | 0 | 447 | 2 | 0 | 2 | 0 | 1 | 1 | 0 | 136 | 28 | 35 | 0 | 0 | 43 |
| 6 | 31 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 68 | 5 | 0 | 189 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 69 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 525 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 354 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 7 | 0 | 86 | 0 | 0 | 0 | 0 | 0 | 213 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 223 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 0 | 0 | 0 |
| 13 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| 14 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 101 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 90 | 0 | 0 | 0 | 1 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 59 | 0 | 0 |
'PBMC4 - contingency_matrix (rows: cellTypist - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ... | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 1 | 0 | 402 | 0 | 0 | 4 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 45 | ... | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 45 | 963 | 322 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 105 | 0 | 0 | 0 | 0 | 3 |
| 5 | 40 | 131 | 724 | 199 | 332 | 741 | 1 | 27 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 209 | 95 |
| 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 3 | 0 | 4 | 2 | 0 | 68 |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 7 | 530 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 55 | 36 | 118 | 147 | 1 | 1 | 0 | 1 |
| 10 | 0 | 1 | 6 | 0 | 83 | 2 | 215 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 24 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | ... | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 93 | 1 | 12 |
| 15 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 90 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 59 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
16 rows × 24 columns
'Initial antibody cell/cluster table:'cluster.ids
3 2280
1 1367
10 1018
9 488
2 351
14 348
4 242
5 224
24 194
26 64
22 43
12 41
Name: count, dtype: int64'PBMC4 - contingency_matrix (rows: antibody - cols: monocle)'| 1 | 2 | 3 | |
|---|---|---|---|
| 1 | 1342 | 5 | 0 |
| 2 | 335 | 5 | 0 |
| 3 | 8 | 2158 | 0 |
| 4 | 241 | 0 | 0 |
| 5 | 16 | 195 | 1 |
| 9 | 480 | 1 | 0 |
| 10 | 13 | 39 | 931 |
| 12 | 38 | 0 | 0 |
| 14 | 343 | 2 | 0 |
| 22 | 42 | 0 | 0 |
| 24 | 193 | 0 | 0 |
| 26 | 63 | 0 | 0 |
'PBMC4 - contingency_matrix (rows: antibody - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ... | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 744 | 0 | 0 | 0 | 195 | 247 | 0 | 8 | 1 | 0 | ... | 139 | 6 | 0 | 0 | 3 | 1 | 2 | 0 | 1 | 0 |
| 2 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 317 | 0 | ... | 2 | 0 | 0 | 0 | 3 | 0 | 3 | 0 | 0 | 0 |
| 3 | 1 | 0 | 573 | 514 | 1 | 0 | 405 | 0 | 0 | 273 | ... | 2 | 0 | 0 | 178 | 26 | 119 | 0 | 42 | 3 | 0 |
| 4 | 3 | 0 | 0 | 0 | 3 | 2 | 0 | 1 | 2 | 0 | ... | 8 | 30 | 174 | 0 | 0 | 0 | 18 | 0 | 0 | 0 |
| 5 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 14 | 0 | ... | 1 | 0 | 0 | 0 | 3 | 11 | 2 | 1 | 0 | 0 |
| 9 | 12 | 0 | 0 | 0 | 1 | 3 | 0 | 288 | 0 | 0 | ... | 2 | 130 | 8 | 0 | 1 | 0 | 36 | 0 | 0 | 0 |
| 10 | 1 | 657 | 1 | 0 | 0 | 0 | 1 | 0 | 6 | 1 | ... | 0 | 0 | 0 | 0 | 40 | 0 | 0 | 3 | 0 | 37 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 0 | ... | 0 | 2 | 0 | 0 | 0 | 0 | 31 | 0 | 0 | 0 |
| 14 | 10 | 0 | 0 | 0 | 205 | 105 | 0 | 0 | 0 | 0 | ... | 23 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| 22 | 6 | 0 | 0 | 0 | 2 | 2 | 0 | 2 | 0 | 0 | ... | 0 | 24 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 24 | 9 | 0 | 0 | 0 | 97 | 75 | 0 | 0 | 1 | 0 | ... | 9 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 26 | 27 | 0 | 0 | 0 | 1 | 12 | 0 | 1 | 0 | 0 | ... | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
12 rows × 22 columns
'PBMC4 - contingency_matrix (rows: antibody - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 454 | 839 | 0 | 1 | 1 | 8 | 1 | 12 | 0 | 0 | 2 | 1 | 0 | 1 | 27 |
| 2 | 0 | 1 | 4 | 1 | 0 | 0 | 0 | 328 | 0 | 0 | 0 | 3 | 1 | 0 | 2 | 0 |
| 3 | 0 | 2 | 0 | 719 | 730 | 476 | 1 | 0 | 2 | 15 | 168 | 14 | 1 | 35 | 3 | 0 |
| 4 | 0 | 4 | 2 | 0 | 0 | 0 | 230 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 | 10 | 1 | 7 | 0 | 19 | 0 | 170 | 0 | 3 | 0 | 1 | 1 | 0 |
| 9 | 0 | 2 | 8 | 0 | 0 | 0 | 175 | 0 | 288 | 0 | 0 | 1 | 7 | 0 | 0 | 0 |
| 10 | 921 | 0 | 3 | 36 | 4 | 1 | 0 | 5 | 0 | 2 | 8 | 0 | 1 | 2 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 38 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 324 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 2 |
| 22 | 0 | 3 | 4 | 0 | 0 | 0 | 27 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 185 | 4 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 26 | 1 | 4 | 58 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC4 - contingency_matrix (rows: antibody - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 853 | 323 | 0 | 0 | 0 | 0 | 9 | 1 | 0 | 5 | 151 | 0 | 0 | 1 | 3 | 0 | 1 | 0 | 0 |
| 2 | 9 | 0 | 0 | 0 | 0 | 0 | 2 | 324 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 1 | 0 |
| 3 | 4 | 1 | 750 | 693 | 0 | 519 | 1 | 0 | 0 | 0 | 1 | 25 | 0 | 113 | 26 | 30 | 3 | 0 | 0 |
| 4 | 1 | 11 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 46 | 2 | 0 | 178 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 1 | 0 | 1 | 2 | 0 | 2 | 1 | 15 | 0 | 0 | 0 | 177 | 0 | 9 | 3 | 1 | 0 | 0 | 0 |
| 9 | 28 | 5 | 0 | 0 | 0 | 0 | 292 | 0 | 0 | 140 | 9 | 0 | 6 | 0 | 1 | 0 | 0 | 0 | 0 |
| 10 | 1 | 0 | 3 | 3 | 598 | 0 | 0 | 4 | 304 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 30 | 37 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 36 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 10 | 326 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| 22 | 12 | 11 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 9 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| 24 | 3 | 186 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 26 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC4 - contingency_matrix (rows: antibody - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ... | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | ... | 1 | 0 | 0 | 0 | 3 | 0 | 9 | 75 | 431 | 824 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 1 | ... | 0 | 0 | 57 | 29 | 102 | 142 | 1 | 0 | 0 | 4 |
| 3 | 0 | 110 | 687 | 195 | 395 | 719 | 26 | 25 | 0 | 0 | ... | 3 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 2 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 1 | 0 | 19 | 0 | 1 | 1 | 191 | 28 |
| 5 | 0 | 9 | 2 | 0 | 1 | 2 | 178 | 3 | 0 | 0 | ... | 0 | 0 | 0 | 4 | 10 | 1 | 0 | 0 | 0 | 1 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 41 | 0 | 292 | 6 | 12 | 129 |
| 10 | 33 | 1 | 4 | 1 | 2 | 2 | 1 | 0 | 24 | 25 | ... | 0 | 0 | 1 | 1 | 1 | 2 | 0 | 0 | 1 | 1 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 36 | 0 | 0 | 1 | 0 | 1 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | ... | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 333 | 9 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 17 | 19 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 187 | 4 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 57 | 1 | 5 |
12 rows × 24 columns
Against cellTypist cluster number
print_clustering_data(tuning = 'celltypist',dataset="PBMC1")'Initial COTAN cluster number:'14'Initial monocle cluster number:'14'Initial scanpy cluster number:'18'Initial scvi-tools cluster number:'17'Initial seurat cluster number:'20'PBMC1 - contingency_matrix (rows: cellTypist - cols: monocle)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 1 | 3 | 1 | 273 | 0 | 0 | 0 | 237 | 227 | 147 | 0 | 6 | 6 | 0 | 77 | 1 | 0 |
| 2 | 66 | 0 | 0 | 0 | 0 | 230 | 228 | 218 | 0 | 0 | 0 | 145 | 25 | 0 | 31 | 0 | 0 | 0 |
| 3 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 36 | 0 | 6 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 73 | 0 |
| 5 | 200 | 0 | 0 | 7 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 65 | 0 | 35 | 0 | 0 | 0 |
| 6 | 0 | 142 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 36 | 0 | 0 | 40 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 1 | 267 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 9 | 2 | 0 | 0 | 0 | 0 | 22 | 15 | 20 | 0 | 0 | 0 | 21 | 0 | 0 | 1 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 37 | 0 | 0 | 131 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 21 | 49 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 27 | 0 | 0 | 179 | 0 | 3 | 2 | 1 | 0 | 0 | 0 | 4 | 14 | 0 | 10 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 |
| 14 | 0 | 155 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 0 |
'PBMC1 - contingency_matrix (rows: cellTypist - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 369 | 243 | 292 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 74 | 0 |
| 2 | 474 | 89 | 0 | 0 | 0 | 258 | 0 | 2 | 0 | 0 | 0 | 111 | 0 | 8 | 0 | 1 | 0 |
| 3 | 0 | 45 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 76 | 0 | 0 |
| 5 | 2 | 260 | 0 | 0 | 0 | 2 | 0 | 42 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 138 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 55 | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 | 263 | 0 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 5 | 2 | 0 | 0 | 0 | 5 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 66 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 78 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 91 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 65 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 11 | 0 | 0 | 0 | 6 | 0 | 143 | 78 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 154 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: cellTypist - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 353 | 261 | 1 | 2 | 0 | 1 | 183 | 0 | 4 | 0 | 152 | 0 | 1 | 19 | 1 | 0 | 0 | 0 | 0 |
| 2 | 43 | 0 | 0 | 278 | 0 | 227 | 187 | 0 | 180 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 16 | 0 | 3 |
| 3 | 4 | 0 | 0 | 9 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 31 | 0 | 0 |
| 4 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 73 | 0 | 0 | 0 | 0 | 0 |
| 5 | 279 | 0 | 0 | 11 | 4 | 9 | 2 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 142 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 55 | 0 | 0 | 0 | 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 152 | 1 | 0 | 0 | 125 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 1 | 0 | 0 | 1 | 1 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 72 | 0 | 0 | 2 |
| 10 | 0 | 0 | 86 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 81 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 65 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 45 | 0 | 0 | 1 | 173 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 17 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 145 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 15 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: cellTypist - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ... | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 351 | 267 | 0 | 229 | 0 | 0 | 0 | 0 | ... | 0 | 3 | 0 | 1 | 88 | 0 | 0 | 40 | 0 | 0 |
| 2 | 423 | 119 | 0 | 0 | 238 | 0 | 154 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 1 | 0 |
| 3 | 1 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 31 | 0 |
| 4 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 74 | 0 | 0 | 0 | 0 |
| 5 | 0 | 258 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 142 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 0 | ... | 0 | 0 | 12 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 151 | 0 | 0 | ... | 0 | 0 | 5 | 123 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 10 | 2 | 0 | 0 | 11 | 0 | 11 | 0 | 1 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 46 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 38 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 130 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | ... | 0 | 0 | 66 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 1 | 3 | 0 | 0 | 4 | 0 | 0 | 0 | 138 | 0 | ... | 0 | 0 | 44 | 0 | 0 | 0 | 2 | 0 | 1 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 27 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 153 | ... | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 |
15 rows × 21 columns
'PBMC1 - contingency_matrix (rows: cellTypist - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 35 | 0 | 11 | 648 | 284 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 807 | 129 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 37 | 2 | 8 |
| 4 | 0 | 73 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 308 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 142 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 1 | 0 | 0 | 56 |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 55 | 4 | 220 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 34 | 1 | 0 | 0 | 45 | 1 |
| 10 | 0 | 1 | 146 | 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 67 | 1 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 175 | 0 | 1 | 5 | 56 |
| 13 | 0 | 1 | 0 | 0 | 0 | 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 152 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
print_clustering_data(tuning = 'celltypist',dataset="PBMC2")'Initial COTAN cluster number:'17'Initial monocle cluster number:'17'Initial scanpy cluster number:'18'Initial scvi-tools cluster number:'20'Initial seurat cluster number:'19'PBMC2 - contingency_matrix (rows: cellTypist - cols: monocle)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 0 | 0 | 1 | 0 | 5 | 0 | 55 | 0 | 0 | 12 | 38 | 6 | 90 | 1 | 0 | 0 | 22 |
| 2 | 0 | 0 | 0 | 0 | 0 | 279 | 0 | 6 | 22 | 0 | 53 | 22 | 24 | 1 | 19 | 0 | 0 | 1 |
| 3 | 0 | 1 | 332 | 1 | 407 | 130 | 338 | 27 | 297 | 36 | 202 | 95 | 99 | 38 | 132 | 0 | 0 | 7 |
| 4 | 577 | 1 | 0 | 6 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 2 | 42 | 0 | 0 | 0 | 75 |
| 5 | 1 | 0 | 0 | 0 | 0 | 15 | 0 | 258 | 0 | 0 | 9 | 9 | 2 | 13 | 0 | 0 | 0 | 9 |
| 6 | 0 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 77 | 0 |
| 7 | 0 | 566 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 142 | 0 | 32 | 4 | 69 | 4 | 11 | 292 | 24 | 59 | 4 | 2 | 31 | 0 | 0 | 0 |
| 9 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 173 | 0 | 0 |
| 10 | 43 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 3 |
| 11 | 0 | 0 | 0 | 228 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 204 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 1 | 3 | 0 | 0 | 0 | 66 | 3 | 3 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 |
'PBMC2 - contingency_matrix (rows: cellTypist - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 1 | 0 | 15 | 2 | 57 | 5 | 0 | 148 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 131 | 200 | 35 | 2 | 2 | 0 | 1 | 55 | 0 | 1 | 0 | 0 | 0 | 0 |
| 3 | 21 | 575 | 490 | 0 | 0 | 0 | 230 | 107 | 248 | 286 | 10 | 2 | 48 | 98 | 0 | 26 | 0 | 0 | 0 | 1 |
| 4 | 0 | 0 | 0 | 0 | 471 | 8 | 0 | 1 | 0 | 0 | 2 | 222 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 253 | 0 | 3 | 52 | 0 | 6 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 88 | 0 | 0 |
| 7 | 0 | 0 | 0 | 464 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 88 | 0 | 0 | 0 |
| 8 | 558 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 112 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 174 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 228 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 204 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 44 | 17 | 1 | 1 | 1 | 2 | 3 | 0 | 1 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC2 - contingency_matrix (rows: cellTypist - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 1 | 0 | 217 | 0 | 0 | 0 | 7 | 0 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| 2 | 348 | 2 | 0 | 0 | 0 | 4 | 7 | 0 | 2 | 3 | 60 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 469 | 4 | 1 | 15 | 1 | 412 | 69 | 356 | 324 | 322 | 165 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 1 | 667 | 1 | 0 | 8 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 23 | 0 | 0 | 0 |
| 5 | 10 | 2 | 0 | 0 | 0 | 0 | 7 | 0 | 1 | 0 | 65 | 0 | 159 | 0 | 72 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 80 | 0 | 0 | 4 | 0 | 0 |
| 7 | 0 | 0 | 556 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 9 |
| 8 | 1 | 0 | 1 | 561 | 0 | 2 | 99 | 1 | 2 | 2 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 173 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 47 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 228 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 204 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 0 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 0 |
| 15 | 61 | 0 | 0 | 0 | 0 | 3 | 2 | 1 | 1 | 3 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 |
'PBMC2 - contingency_matrix (rows: cellTypist - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 1 | 0 | 1 | 41 | 178 | 0 | 0 | 1 | 0 | 0 | 0 |
| 2 | 389 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 10 | 3 | 0 | 23 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 259 | 0 | 578 | 568 | 15 | 0 | 1 | 290 | 188 | 0 | 2 | 169 | 62 | 8 | 0 | 0 | 2 | 0 | 0 | 0 |
| 4 | 0 | 635 | 0 | 0 | 0 | 0 | 7 | 0 | 2 | 0 | 0 | 0 | 0 | 3 | 0 | 1 | 59 | 0 | 0 | 0 |
| 5 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 268 | 0 | 32 | 1 | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 82 | 0 | 0 |
| 7 | 0 | 0 | 0 | 0 | 0 | 305 | 0 | 0 | 0 | 0 | 262 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 1 | 0 | 8 | 0 | 541 | 0 | 0 | 0 | 1 | 0 | 0 | 3 | 119 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 172 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 83 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 145 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 201 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 |
| 15 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 76 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 |
'PBMC2 - contingency_matrix (rows: cellTypist - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 3 | 4 | 220 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 416 | 9 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 2 | 1186 | 847 | 31 | 72 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 4 | 639 | 56 | 0 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 5 | 1 |
| 5 | 0 | 0 | 0 | 39 | 0 | 7 | 270 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 86 | 0 | 7 | 0 | 0 | 0 | 0 | 0 |
| 7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 88 | 300 | 174 | 0 | 0 | 3 | 1 |
| 8 | 0 | 0 | 568 | 9 | 1 | 96 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 154 | 24 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 |
| 10 | 1 | 51 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 145 | 69 | 14 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 22 | 180 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 1 |
| 15 | 0 | 1 | 0 | 6 | 72 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 0 |
print_clustering_data(tuning = 'celltypist',dataset="PBMC3")'Initial COTAN cluster number:'21'Initial monocle cluster number:'21'Initial scanpy cluster number:'17'Initial scvi-tools cluster number:'18'Initial seurat cluster number:'20'PBMC3 - contingency_matrix (rows: cellTypist - cols: monocle)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 860 | 0 | 0 | 0 | 38 | 559 | 313 | 338 | 231 | 324 | 158 | 0 | 195 | 0 | 0 | 5 | 0 |
| 2 | 0 | 1041 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 390 | 0 | 2 | 7 | 0 | 31 |
| 3 | 0 | 0 | 0 | 654 | 0 | 0 | 2 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 |
| 4 | 400 | 0 | 0 | 0 | 12 | 309 | 29 | 47 | 12 | 157 | 78 | 0 | 56 | 0 | 0 | 0 | 0 |
| 5 | 0 | 22 | 1036 | 33 | 22 | 0 | 0 | 0 | 0 | 0 | 3 | 2 | 1 | 0 | 2 | 121 | 0 |
| 6 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 144 | 0 | 0 |
| 7 | 0 | 0 | 0 | 0 | 134 | 1 | 247 | 305 | 248 | 0 | 135 | 1 | 42 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 3 | 0 | 435 | 0 | 3 | 7 | 0 | 0 | 12 | 0 | 20 | 0 | 0 | 4 | 0 |
| 9 | 0 | 0 | 0 | 396 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 73 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 328 | 2 | 0 | 1 |
| 11 | 0 | 0 | 8 | 0 | 277 | 2 | 21 | 26 | 1 | 0 | 47 | 0 | 46 | 0 | 0 | 2 | 0 |
| 12 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 11 | 0 | 0 | 0 | 0 | 8 | 157 | 9 | 19 | 1 | 7 | 0 | 21 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 79 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 19 | 0 |
| 15 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 57 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 |
'PBMC3 - contingency_matrix (rows: cellTypist - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1670 | 0 | 0 | 212 | 0 | 32 | 234 | 621 | 23 | 228 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 1228 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 202 | 0 | 0 | 5 | 0 | 35 | 0 |
| 3 | 0 | 0 | 0 | 0 | 543 | 0 | 5 | 0 | 0 | 1 | 0 | 0 | 0 | 111 | 0 | 0 | 2 | 0 |
| 4 | 29 | 0 | 0 | 0 | 0 | 811 | 192 | 5 | 4 | 59 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 0 | 0 | 1028 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 29 | 145 | 33 | 0 | 0 | 0 | 0 |
| 6 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 149 | 0 | 0 | 0 |
| 7 | 1 | 0 | 0 | 702 | 0 | 0 | 230 | 99 | 7 | 73 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 3 | 0 | 0 | 0 | 47 | 0 | 433 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 395 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| 10 | 0 | 94 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 311 | 1 | 0 | 0 | 1 | 0 | 1 | 0 |
| 11 | 0 | 0 | 2 | 0 | 0 | 0 | 116 | 0 | 290 | 21 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 |
| 13 | 16 | 0 | 0 | 44 | 0 | 0 | 10 | 55 | 1 | 107 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 11 | 0 | 0 | 0 | 4 | 0 | 5 | 1 | 0 | 0 | 90 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 20 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 57 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 |
'PBMC3 - contingency_matrix (rows: cellTypist - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 657 | 1415 | 2 | 2 | 43 | 614 | 91 | 5 | 0 | 64 | 0 | 14 | 49 | 0 | 0 | 32 | 31 | 0 | 0 |
| 2 | 1463 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 4 | 0 | 1 | 0 | 654 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 6 | 13 | 1 | 0 | 856 | 4 | 168 | 0 | 0 | 0 | 0 | 0 | 52 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 29 | 0 | 0 | 1172 | 32 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 1 | 0 |
| 6 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 147 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 1 | 827 | 5 | 0 | 0 | 1 | 187 | 35 | 22 | 0 | 0 | 0 | 24 | 8 | 0 | 0 | 1 | 2 | 0 | 0 |
| 8 | 0 | 30 | 0 | 2 | 0 | 1 | 3 | 12 | 305 | 0 | 131 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 396 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 83 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 324 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 15 | 0 | 2 | 1 | 3 | 1 | 399 | 3 | 0 | 5 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 |
| 13 | 0 | 49 | 21 | 0 | 0 | 0 | 22 | 6 | 0 | 0 | 0 | 0 | 131 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 80 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 20 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 52 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
'PBMC3 - contingency_matrix (rows: cellTypist - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1744 | 412 | 0 | 0 | 681 | 20 | 161 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 1011 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 227 | 0 | 231 | 3 | 0 | 0 | 0 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 535 | 0 | 2 | 0 | 1 | 2 | 0 | 0 | 120 | 0 | 0 |
| 4 | 16 | 6 | 0 | 0 | 3 | 886 | 188 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 5 | 0 | 1 | 4 | 1043 | 0 | 0 | 2 | 1 | 1 | 0 | 0 | 24 | 136 | 0 | 0 | 30 | 0 | 0 |
| 6 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 149 | 0 | 0 | 0 |
| 7 | 0 | 995 | 0 | 0 | 93 | 0 | 18 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 18 | 0 | 3 | 0 | 0 | 8 | 0 | 454 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 57 | 0 | 336 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| 10 | 0 | 0 | 89 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 314 | 1 | 0 | 4 | 0 | 0 | 0 | 0 |
| 11 | 0 | 14 | 0 | 0 | 0 | 2 | 411 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 13 | 13 | 8 | 0 | 0 | 205 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 109 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 20 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 52 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 |
'PBMC3 - contingency_matrix (rows: cellTypist - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ... | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 205 | 1805 | 252 | 548 | 154 | 55 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | ... | 177 | 274 | 220 | 795 | 0 | 0 | 0 | 1 | 0 | 0 |
| 3 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | ... | 0 | 0 | 0 | 1 | 67 | 107 | 225 | 256 | 0 | 0 |
| 4 | 89 | 823 | 9 | 6 | 20 | 153 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 1 | 0 | 1 | 1 | 0 | 1 | 1029 | 138 | 30 | 0 | ... | 2 | 1 | 1 | 5 | 3 | 26 | 3 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 146 | ... | 0 | 0 | 8 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 26 | 0 | 977 | 61 | 27 | 21 | 0 | 1 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 9 | 1 | 0 | 469 | 3 | 2 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 2 | 379 | 15 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 40 | 5 | 119 | 27 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 9 | 1 | 7 | 1 | 8 | 357 | 1 | 14 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 32 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 |
| 13 | 4 | 5 | 4 | 204 | 6 | 0 | 0 | 10 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 1 | 0 | 1 | 0 | 0 | 0 | 92 | 17 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 52 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 |
19 rows × 21 columns
print_clustering_data(tuning = 'celltypist',dataset="PBMC4")'Initial COTAN cluster number:'19'Initial monocle cluster number:'21'Initial scanpy cluster number:'16'Initial scvi-tools cluster number:'18'Initial seurat cluster number:'18'PBMC4 - contingency_matrix (rows: cellTypist - cols: monocle)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 12 | 0 | 385 | 1 | 0 | 0 | 0 | 0 | 0 |
| 2 | 756 | 2 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 3 | 3 | 0 | 0 | 0 | 0 | 41 |
| 3 | 0 | 707 | 149 | 0 | 0 | 0 | 379 | 0 | 3 | 0 | 93 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 98 | 0 | 8 | 1 | 0 | 0 | 0 | 0 | 0 |
| 5 | 5 | 2 | 2 | 648 | 642 | 406 | 0 | 1 | 422 | 3 | 2 | 3 | 35 | 0 | 29 | 0 |
| 6 | 0 | 196 | 83 | 0 | 0 | 0 | 19 | 1 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 0 | 51 | 0 | 0 | 0 | 0 | 9 | 0 | 15 | 2 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 2 | 417 | 0 | 0 | 0 | 77 | 0 | 0 | 5 | 37 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 351 | 1 | 3 | 4 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 2 | 3 | 76 | 0 | 0 | 5 | 0 | 0 | 219 | 2 | 0 | 0 | 0 |
| 11 | 220 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 2 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 0 |
| 13 | 1 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | 0 |
| 14 | 0 | 4 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 97 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 11 | 5 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 72 | 3 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 59 | 0 | 0 |
'PBMC4 - contingency_matrix (rows: cellTypist - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 10 | 0 | 0 | 0 | 0 | 385 | 5 | 1 | 0 | 0 | 0 | 0 | 1 | 5 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 673 | 0 | 0 | 0 | 0 | 1 | 134 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 346 | 0 | 0 | 509 | 387 | 1 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 78 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 7 | 10 | 0 | 0 | 0 | 0 | 0 | 85 | 0 | 0 | 0 |
| 5 | 928 | 1 | 822 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 181 | 78 | 142 | 0 | 0 | 47 | 0 |
| 6 | 0 | 7 | 0 | 0 | 5 | 2 | 0 | 286 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 |
| 7 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 67 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 |
| 8 | 0 | 483 | 0 | 0 | 8 | 43 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 9 | 0 | 1 | 0 | 0 | 0 | 0 | 5 | 0 | 348 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 |
| 10 | 74 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 216 | 10 | 2 | 2 | 0 | 0 | 1 | 0 |
| 11 | 0 | 0 | 0 | 46 | 0 | 0 | 0 | 0 | 0 | 177 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 0 |
| 13 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 68 | 0 | 0 | 1 | 22 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 89 | 2 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 59 |
'PBMC4 - contingency_matrix (rows: cellTypist - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 313 | 0 | 0 | 1 | 0 | 0 | 88 | 2 | 0 | 0 |
| 2 | 1 | 1 | 0 | 0 | 687 | 0 | 1 | 1 | 10 | 0 | 104 | 2 | 0 | 0 | 0 | 1 | 0 | 0 |
| 3 | 439 | 840 | 1 | 0 | 0 | 0 | 0 | 21 | 0 | 0 | 0 | 0 | 30 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 1 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 1 | 1 | 776 | 699 | 2 | 508 | 15 | 0 | 0 | 65 | 2 | 1 | 0 | 24 | 2 | 84 | 0 | 20 |
| 6 | 10 | 8 | 0 | 0 | 0 | 0 | 0 | 287 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| 7 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 33 | 5 | 0 | 0 | 34 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 507 | 15 | 0 | 0 | 0 | 0 | 1 | 3 | 2 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 358 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 13 | 1 | 0 | 27 | 0 | 0 | 0 | 265 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 219 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 |
| 13 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 16 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 81 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 89 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 57 | 0 |
'PBMC4 - contingency_matrix (rows: cellTypist - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 0 | 0 | 0 | 0 | 0 | 398 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 632 | 0 | 1 | 0 | 122 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 52 | 0 |
| 3 | 361 | 843 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 122 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 107 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 2 | 0 | 771 | 732 | 0 | 447 | 2 | 0 | 2 | 0 | 1 | 1 | 0 | 136 | 28 | 35 | 0 | 0 | 43 |
| 6 | 31 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 68 | 5 | 0 | 189 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 69 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 525 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 354 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 7 | 0 | 86 | 0 | 0 | 0 | 0 | 0 | 213 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 223 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 0 | 0 | 0 |
| 13 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| 14 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 101 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 90 | 0 | 0 | 0 | 1 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 59 | 0 | 0 |
'PBMC4 - contingency_matrix (rows: cellTypist - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 402 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 45 | 2 | 24 | 320 | 414 | 0 | 0 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 367 | 963 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 105 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 40 | 131 | 724 | 531 | 741 | 1 | 27 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 99 | 209 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 4 | 70 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 530 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 91 | 265 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 1 | 6 | 83 | 2 | 215 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 24 | 141 | 58 | 1 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 8 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 105 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 1 | 0 | 1 | 1 | 0 | 0 | 90 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 59 | 0 |
Against antibody cluster number
print_clustering_data(tuning = 'antibody',dataset="PBMC1")'Initial COTAN cluster number:'11'Initial monocle cluster number:'11'Initial scanpy cluster number:'9'Initial scvi-tools cluster number:'11'Initial seurat cluster number:'10'Initial antibody cell/cluster table:'cluster.ids
7 1338
8 876
3 748
4 341
9 331
5 211
1 202
6 131
2 62
10 51
12 16
Name: count, dtype: int64'PBMC1 - contingency_matrix (rows: antibody - cols: monocle)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 94 | 0 | 17 | 50 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 4 | 0 | 0 | 3 | 0 | 38 | 1 | 0 |
| 3 | 35 | 0 | 500 | 65 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 1 | 0 | 262 | 0 | 0 | 0 | 0 | 0 |
| 5 | 2 | 0 | 29 | 127 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 58 | 26 |
| 7 | 2 | 731 | 0 | 4 | 0 | 275 | 95 | 17 | 1 |
| 8 | 776 | 0 | 30 | 6 | 1 | 0 | 0 | 0 | 1 |
| 9 | 0 | 0 | 0 | 1 | 294 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 44 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 22 | 0 | 7 | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 72 |
| 2 | 0 | 0 | 0 | 0 | 5 | 0 | 2 | 1 | 0 | 38 | 0 |
| 3 | 67 | 0 | 408 | 125 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 1 | 0 | 13 | 0 | 249 | 0 | 0 | 0 | 0 | 0 |
| 5 | 2 | 0 | 10 | 146 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 82 | 1 | 0 |
| 7 | 2 | 702 | 1 | 0 | 348 | 0 | 0 | 0 | 20 | 52 | 0 |
| 8 | 780 | 0 | 21 | 6 | 0 | 1 | 1 | 0 | 1 | 0 | 4 |
| 9 | 0 | 0 | 0 | 1 | 0 | 1 | 156 | 137 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 37 | 0 | 7 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 28 | 0 | 0 | 22 | 0 | 1 | 36 | 0 | 74 | 0 |
| 2 | 0 | 3 | 42 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 3 | 123 | 0 | 0 | 434 | 0 | 0 | 43 | 0 | 0 | 0 |
| 4 | 0 | 1 | 0 | 0 | 0 | 259 | 3 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 | 17 | 0 | 0 | 138 | 0 | 3 | 0 |
| 6 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 56 | 3 | 26 |
| 7 | 1 | 644 | 451 | 0 | 0 | 1 | 0 | 27 | 0 | 1 |
| 8 | 786 | 0 | 0 | 19 | 1 | 2 | 3 | 0 | 2 | 1 |
| 9 | 0 | 0 | 0 | 0 | 294 | 1 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 1 | 0 | 9 | 34 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 93 | 14 | 0 | 0 | 0 | 54 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 1 | 4 | 0 | 0 | 1 | 1 | 39 | 0 | 0 |
| 3 | 22 | 536 | 0 | 0 | 0 | 42 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 1 | 0 | 261 | 1 | 0 | 0 | 0 | 0 | 0 |
| 5 | 1 | 18 | 0 | 0 | 0 | 139 | 0 | 0 | 0 | 0 | 0 |
| 6 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 56 | 25 |
| 7 | 2 | 1 | 524 | 488 | 2 | 0 | 0 | 0 | 89 | 18 | 1 |
| 8 | 766 | 40 | 0 | 0 | 1 | 5 | 1 | 0 | 0 | 0 | 1 |
| 9 | 0 | 0 | 0 | 0 | 2 | 0 | 151 | 142 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 8 | 36 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 8 | 2 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 36 | 54 | 12 | 59 |
| 2 | 1 | 1 | 40 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 39 | 528 | 33 |
| 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 214 | 0 | 0 |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 135 | 22 | 0 |
| 6 | 0 | 56 | 1 | 1 | 1 | 25 | 0 | 3 | 0 | 0 | 0 |
| 7 | 33 | 17 | 118 | 668 | 284 | 1 | 0 | 0 | 1 | 1 | 2 |
| 8 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 9 | 4 | 34 | 765 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 293 | 0 | 2 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 40 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 6 | 0 | 0 |
print_clustering_data(tuning = 'antibody',dataset="PBMC1")'Initial COTAN cluster number:'11'Initial monocle cluster number:'11'Initial scanpy cluster number:'9'Initial scvi-tools cluster number:'11'Initial seurat cluster number:'10'Initial antibody cell/cluster table:'cluster.ids
7 1338
8 876
3 748
4 341
9 331
5 211
1 202
6 131
2 62
10 51
12 16
Name: count, dtype: int64'PBMC1 - contingency_matrix (rows: antibody - cols: monocle)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 94 | 0 | 17 | 50 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 4 | 0 | 0 | 3 | 0 | 38 | 1 | 0 |
| 3 | 35 | 0 | 500 | 65 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 1 | 0 | 262 | 0 | 0 | 0 | 0 | 0 |
| 5 | 2 | 0 | 29 | 127 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 58 | 26 |
| 7 | 2 | 731 | 0 | 4 | 0 | 275 | 95 | 17 | 1 |
| 8 | 776 | 0 | 30 | 6 | 1 | 0 | 0 | 0 | 1 |
| 9 | 0 | 0 | 0 | 1 | 294 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 44 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 22 | 0 | 7 | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 72 |
| 2 | 0 | 0 | 0 | 0 | 5 | 0 | 2 | 1 | 0 | 38 | 0 |
| 3 | 67 | 0 | 408 | 125 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 1 | 0 | 13 | 0 | 249 | 0 | 0 | 0 | 0 | 0 |
| 5 | 2 | 0 | 10 | 146 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 82 | 1 | 0 |
| 7 | 2 | 702 | 1 | 0 | 348 | 0 | 0 | 0 | 20 | 52 | 0 |
| 8 | 780 | 0 | 21 | 6 | 0 | 1 | 1 | 0 | 1 | 0 | 4 |
| 9 | 0 | 0 | 0 | 1 | 0 | 1 | 156 | 137 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 37 | 0 | 7 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 28 | 0 | 0 | 22 | 0 | 1 | 36 | 0 | 74 | 0 |
| 2 | 0 | 3 | 42 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 3 | 123 | 0 | 0 | 434 | 0 | 0 | 43 | 0 | 0 | 0 |
| 4 | 0 | 1 | 0 | 0 | 0 | 259 | 3 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 | 17 | 0 | 0 | 138 | 0 | 3 | 0 |
| 6 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 56 | 3 | 26 |
| 7 | 1 | 644 | 451 | 0 | 0 | 1 | 0 | 27 | 0 | 1 |
| 8 | 786 | 0 | 0 | 19 | 1 | 2 | 3 | 0 | 2 | 1 |
| 9 | 0 | 0 | 0 | 0 | 294 | 1 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 1 | 0 | 9 | 34 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 93 | 14 | 0 | 0 | 0 | 54 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 1 | 4 | 0 | 0 | 1 | 1 | 39 | 0 | 0 |
| 3 | 22 | 536 | 0 | 0 | 0 | 42 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 1 | 0 | 261 | 1 | 0 | 0 | 0 | 0 | 0 |
| 5 | 1 | 18 | 0 | 0 | 0 | 139 | 0 | 0 | 0 | 0 | 0 |
| 6 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 56 | 25 |
| 7 | 2 | 1 | 524 | 488 | 2 | 0 | 0 | 0 | 89 | 18 | 1 |
| 8 | 766 | 40 | 0 | 0 | 1 | 5 | 1 | 0 | 0 | 0 | 1 |
| 9 | 0 | 0 | 0 | 0 | 2 | 0 | 151 | 142 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 8 | 36 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 8 | 2 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 36 | 54 | 12 | 59 |
| 2 | 1 | 1 | 40 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 39 | 528 | 33 |
| 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 214 | 0 | 0 |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 135 | 22 | 0 |
| 6 | 0 | 56 | 1 | 1 | 1 | 25 | 0 | 3 | 0 | 0 | 0 |
| 7 | 33 | 17 | 118 | 668 | 284 | 1 | 0 | 0 | 1 | 1 | 2 |
| 8 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 9 | 4 | 34 | 765 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 293 | 0 | 2 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 40 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 6 | 0 | 0 |
print_clustering_data(tuning = 'antibody',dataset="PBMC1")'Initial COTAN cluster number:'11'Initial monocle cluster number:'11'Initial scanpy cluster number:'9'Initial scvi-tools cluster number:'11'Initial seurat cluster number:'10'Initial antibody cell/cluster table:'cluster.ids
7 1338
8 876
3 748
4 341
9 331
5 211
1 202
6 131
2 62
10 51
12 16
Name: count, dtype: int64'PBMC1 - contingency_matrix (rows: antibody - cols: monocle)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 94 | 0 | 17 | 50 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 4 | 0 | 0 | 3 | 0 | 38 | 1 | 0 |
| 3 | 35 | 0 | 500 | 65 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 1 | 0 | 262 | 0 | 0 | 0 | 0 | 0 |
| 5 | 2 | 0 | 29 | 127 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 58 | 26 |
| 7 | 2 | 731 | 0 | 4 | 0 | 275 | 95 | 17 | 1 |
| 8 | 776 | 0 | 30 | 6 | 1 | 0 | 0 | 0 | 1 |
| 9 | 0 | 0 | 0 | 1 | 294 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 44 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: scanpy)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 22 | 0 | 7 | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 72 |
| 2 | 0 | 0 | 0 | 0 | 5 | 0 | 2 | 1 | 0 | 38 | 0 |
| 3 | 67 | 0 | 408 | 125 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 1 | 0 | 13 | 0 | 249 | 0 | 0 | 0 | 0 | 0 |
| 5 | 2 | 0 | 10 | 146 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 82 | 1 | 0 |
| 7 | 2 | 702 | 1 | 0 | 348 | 0 | 0 | 0 | 20 | 52 | 0 |
| 8 | 780 | 0 | 21 | 6 | 0 | 1 | 1 | 0 | 1 | 0 | 4 |
| 9 | 0 | 0 | 0 | 1 | 0 | 1 | 156 | 137 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 37 | 0 | 7 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: scvi-tools)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 28 | 0 | 0 | 22 | 0 | 1 | 36 | 0 | 74 | 0 |
| 2 | 0 | 3 | 42 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 3 | 123 | 0 | 0 | 434 | 0 | 0 | 43 | 0 | 0 | 0 |
| 4 | 0 | 1 | 0 | 0 | 0 | 259 | 3 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 | 17 | 0 | 0 | 138 | 0 | 3 | 0 |
| 6 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 56 | 3 | 26 |
| 7 | 1 | 644 | 451 | 0 | 0 | 1 | 0 | 27 | 0 | 1 |
| 8 | 786 | 0 | 0 | 19 | 1 | 2 | 3 | 0 | 2 | 1 |
| 9 | 0 | 0 | 0 | 0 | 294 | 1 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 1 | 0 | 9 | 34 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: seurat)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 93 | 14 | 0 | 0 | 0 | 54 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 1 | 4 | 0 | 0 | 1 | 1 | 39 | 0 | 0 |
| 3 | 22 | 536 | 0 | 0 | 0 | 42 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 1 | 0 | 261 | 1 | 0 | 0 | 0 | 0 | 0 |
| 5 | 1 | 18 | 0 | 0 | 0 | 139 | 0 | 0 | 0 | 0 | 0 |
| 6 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 56 | 25 |
| 7 | 2 | 1 | 524 | 488 | 2 | 0 | 0 | 0 | 89 | 18 | 1 |
| 8 | 766 | 40 | 0 | 0 | 1 | 5 | 1 | 0 | 0 | 0 | 1 |
| 9 | 0 | 0 | 0 | 0 | 2 | 0 | 151 | 142 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 8 | 36 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 8 | 2 | 0 | 0 | 0 | 0 | 0 |
'PBMC1 - contingency_matrix (rows: antibody - cols: COTAN)'| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 36 | 54 | 12 | 59 |
| 2 | 1 | 1 | 40 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 39 | 528 | 33 |
| 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 214 | 0 | 0 |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 135 | 22 | 0 |
| 6 | 0 | 56 | 1 | 1 | 1 | 25 | 0 | 3 | 0 | 0 | 0 |
| 7 | 33 | 17 | 118 | 668 | 284 | 1 | 0 | 0 | 1 | 1 | 2 |
| 8 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 9 | 4 | 34 | 765 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 293 | 0 | 2 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 40 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 6 | 0 | 0 |
Default parameters
print_scores(tuning = 'default',dataset="PBMC1")'PBMC1 - number of clusters'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 3 | 18 | 13 | 11 | 14 |
'PBMC1 - Silhuette (higher is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 0.100032 | 0.043761 | 0.065534 | 0.148254 | 0.13383 |
'PBMC1 - davies_bouldin (lower is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 0.893661 | 2.47358 | 2.574291 | 1.392309 | 1.728304 |
'PBMC1 - default labels'| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.578257 | 0.384609 | 0.410140 | 0.979930 | 0.602512 |
| scanpy | 0.721042 | 0.404607 | 0.824980 | 0.640363 | 0.508176 |
| scvi-tools | 0.776232 | 0.599664 | 0.809790 | 0.745344 | 0.666244 |
| seurat | 0.793630 | 0.649593 | 0.784165 | 0.803327 | 0.705921 |
| COTAN | 0.787289 | 0.670392 | 0.803876 | 0.771373 | 0.723485 |
| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.622344 | 0.439255 | 0.458549 | 0.968180 | 0.645589 |
| scanpy | 0.662480 | 0.389320 | 0.814398 | 0.558329 | 0.511739 |
| scvi-tools | 0.718265 | 0.557951 | 0.800919 | 0.651075 | 0.643426 |
| seurat | 0.747924 | 0.647338 | 0.787235 | 0.712353 | 0.712527 |
| COTAN | 0.727398 | 0.637277 | 0.792854 | 0.671926 | 0.705427 |
print_scores(tuning = 'default',dataset="PBMC2")'PBMC2 - number of clusters'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 2 | 18 | 20 | 14 | 17 |
'PBMC2 - Silhuette (higher is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 0.224441 | 0.059225 | 0.000832 | 0.111509 | 0.12581 |
'PBMC2 - davies_bouldin (lower is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 1.866736 | 2.073818 | 3.935702 | 1.630557 | 1.855098 |
'PBMC2 - default labels'| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.393166 | 0.207180 | 0.245998 | 0.978626 | 0.521364 |
| scanpy | 0.718820 | 0.457213 | 0.804000 | 0.649960 | 0.556684 |
| scvi-tools | 0.699788 | 0.424696 | 0.785920 | 0.630670 | 0.525031 |
| seurat | 0.775988 | 0.562430 | 0.819560 | 0.736815 | 0.640108 |
| COTAN | 0.729355 | 0.472800 | 0.745550 | 0.713848 | 0.562480 |
| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.261544 | 0.093122 | 0.154169 | 0.861687 | 0.457589 |
| scanpy | 0.685644 | 0.523244 | 0.773965 | 0.615415 | 0.607208 |
| scvi-tools | 0.654391 | 0.485789 | 0.750936 | 0.579842 | 0.573975 |
| seurat | 0.748480 | 0.679377 | 0.793713 | 0.708124 | 0.734394 |
| COTAN | 0.703575 | 0.633882 | 0.697485 | 0.709773 | 0.704954 |
print_scores(tuning = 'default',dataset="PBMC3")'PBMC3 - number of clusters'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 3 | 22 | 17 | 18 | 32 |
'PBMC3 - Silhuette (higher is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 0.171584 | 0.007616 | 0.055559 | 0.111834 | 0.092445 |
'PBMC3 - davies_bouldin (lower is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 0.999779 | 2.376915 | 2.058343 | 1.698551 | 2.281481 |
'PBMC3 - default labels'| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.500696 | 0.233560 | 0.338609 | 0.960446 | 0.500077 |
| scanpy | 0.685919 | 0.462762 | 0.763719 | 0.622505 | 0.541286 |
| scvi-tools | 0.738418 | 0.579677 | 0.757237 | 0.720511 | 0.635237 |
| seurat | 0.770512 | 0.585110 | 0.821173 | 0.725738 | 0.644073 |
| COTAN | 0.723833 | 0.527470 | 0.849029 | 0.630815 | 0.609217 |
| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.469679 | 0.196545 | 0.316293 | 0.911906 | 0.473204 |
| scanpy | 0.675464 | 0.545251 | 0.756553 | 0.610075 | 0.612243 |
| scvi-tools | 0.721883 | 0.667226 | 0.738229 | 0.706244 | 0.710211 |
| seurat | 0.749240 | 0.667468 | 0.798924 | 0.705374 | 0.712865 |
| COTAN | 0.677769 | 0.535850 | 0.798894 | 0.588538 | 0.606241 |
print_scores(tuning = 'default',dataset="PBMC4")'PBMC4 - number of clusters'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 3 | 22 | 16 | 19 | 24 |
'PBMC4 - Silhuette (higher is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 0.081765 | 0.050853 | 0.061618 | 0.112255 | 0.103418 |
'PBMC4 - davies_bouldin (lower is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 0.954689 | 2.100956 | 2.087707 | 1.442075 | 1.823095 |
'PBMC4 - default labels'| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.617025 | 0.470070 | 0.453383 | 0.965513 | 0.647279 |
| scanpy | 0.701228 | 0.380357 | 0.819943 | 0.612541 | 0.487560 |
| scvi-tools | 0.739299 | 0.504966 | 0.788229 | 0.696088 | 0.584900 |
| seurat | 0.760207 | 0.494746 | 0.847372 | 0.689301 | 0.583823 |
| COTAN | 0.716555 | 0.435917 | 0.808134 | 0.643618 | 0.526063 |
| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.636102 | 0.525318 | 0.482349 | 0.933740 | 0.689338 |
| scanpy | 0.642165 | 0.368291 | 0.789153 | 0.541335 | 0.491476 |
| scvi-tools | 0.698167 | 0.482058 | 0.770861 | 0.638003 | 0.577373 |
| seurat | 0.691539 | 0.444871 | 0.803112 | 0.607186 | 0.550863 |
| COTAN | 0.625477 | 0.351843 | 0.733504 | 0.545185 | 0.461853 |
Matching cellTypist clusters number
print_scores(tuning = 'celltypist',dataset="PBMC1")'PBMC1 - number of clusters'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 18 | 17 | 20 | 21 | 14 |
'PBMC1 - Silhuette (higher is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | celltypist | |
|---|---|---|---|---|---|---|
| 0 | 0.005501 | 0.050843 | 0.063871 | 0.088209 | 0.13383 | 0.090989 |
'PBMC1 - davies_bouldin (lower is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | celltypist | |
|---|---|---|---|---|---|---|
| 0 | 2.83457 | 2.286672 | 2.797123 | 1.984746 | 1.728304 | 1.491801 |
'PBMC1 - matching celltypist labels'| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.658065 | 0.341945 | 0.757164 | 0.581903 | 0.448601 |
| scanpy | 0.735830 | 0.459735 | 0.822412 | 0.665742 | 0.553086 |
| scvi-tools | 0.699950 | 0.375082 | 0.808964 | 0.616828 | 0.479652 |
| seurat | 0.730468 | 0.423069 | 0.849278 | 0.640820 | 0.527386 |
| COTAN | 0.787289 | 0.670392 | 0.803876 | 0.771373 | 0.723485 |
print_scores(tuning = 'celltypist',dataset="PBMC2")'PBMC2 - number of clusters'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 18 | 20 | 19 | 20 | 17 |
'PBMC2 - Silhuette (higher is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | celltypist | |
|---|---|---|---|---|---|---|
| 0 | -0.03374 | 0.025173 | 0.030773 | 0.060901 | 0.12581 | 0.131097 |
'PBMC2 - davies_bouldin (lower is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | celltypist | |
|---|---|---|---|---|---|---|
| 0 | 3.167255 | 2.376025 | 3.431314 | 2.147939 | 1.855098 | 1.231923 |
'PBMC2 - matching celltypist labels'| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.605942 | 0.312112 | 0.699644 | 0.534375 | 0.425421 |
| scanpy | 0.697287 | 0.377675 | 0.809335 | 0.612491 | 0.492889 |
| scvi-tools | 0.709450 | 0.398779 | 0.791930 | 0.642531 | 0.500807 |
| seurat | 0.738307 | 0.418942 | 0.850535 | 0.652244 | 0.529176 |
| COTAN | 0.729355 | 0.472800 | 0.745550 | 0.713848 | 0.562480 |
print_scores(tuning = 'celltypist',dataset="PBMC3")'PBMC3 - number of clusters'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 17 | 18 | 20 | 18 | 21 |
'PBMC3 - Silhuette (higher is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | celltypist | |
|---|---|---|---|---|---|---|
| 0 | -0.039293 | 0.040964 | 0.003634 | 0.112264 | 0.047585 | 0.130032 |
'PBMC3 - davies_bouldin (lower is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | celltypist | |
|---|---|---|---|---|---|---|
| 0 | 3.778924 | 1.889045 | 2.221019 | 1.698481 | 2.344713 | 1.140713 |
'PBMC3 - matching celltypist labels'| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.593459 | 0.350206 | 0.643738 | 0.550465 | 0.432058 |
| scanpy | 0.712344 | 0.545918 | 0.758076 | 0.671816 | 0.609354 |
| scvi-tools | 0.735127 | 0.565025 | 0.767444 | 0.705423 | 0.623277 |
| seurat | 0.771047 | 0.586941 | 0.821567 | 0.726381 | 0.645653 |
| COTAN | 0.677393 | 0.470051 | 0.717462 | 0.641563 | 0.538494 |
print_scores(tuning = 'celltypist',dataset="PBMC4")'PBMC4 - number of clusters'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 16 | 18 | 18 | 19 | 19 |
'PBMC4 - Silhuette (higher is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | celltypist | |
|---|---|---|---|---|---|---|
| 0 | 0.022385 | 0.048061 | 0.107921 | 0.111704 | 0.098387 | 0.081772 |
'PBMC4 - davies_bouldin (lower is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | celltypist | |
|---|---|---|---|---|---|---|
| 0 | 2.007518 | 1.898663 | 1.612564 | 1.442525 | 1.969481 | 1.194603 |
'PBMC4 - matching celltypist labels'| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.686019 | 0.421166 | 0.747399 | 0.633956 | 0.512728 |
| scanpy | 0.730100 | 0.473433 | 0.810168 | 0.664434 | 0.562407 |
| scvi-tools | 0.752718 | 0.500477 | 0.831022 | 0.687899 | 0.587099 |
| seurat | 0.759495 | 0.492528 | 0.846776 | 0.688525 | 0.581840 |
| COTAN | 0.724052 | 0.447103 | 0.782942 | 0.673402 | 0.532939 |
Matching antibody clusters number
print_scores(tuning = 'antibody',dataset="PBMC1")'PBMC1 - number of clusters'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 9 | 11 | 10 | 11 | 11 |
'PBMC1 - Silhuette (higher is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | antibody | |
|---|---|---|---|---|---|---|
| 0 | 0.099643 | 0.073115 | 0.069687 | 0.150193 | 0.090531 | 0.047275 |
'PBMC1 - davies_bouldin (lower is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | antibody | |
|---|---|---|---|---|---|---|
| 0 | 1.498221 | 1.689467 | 1.560514 | 1.392406 | 2.067883 | 1.794015 |
'PBMC1 - matching antibody labels'| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.724319 | 0.641765 | 0.727281 | 0.721381 | 0.707943 |
| scanpy | 0.746106 | 0.652841 | 0.792721 | 0.704669 | 0.717629 |
| scvi-tools | 0.757587 | 0.658079 | 0.782127 | 0.734540 | 0.721084 |
| seurat | 0.749425 | 0.642110 | 0.790860 | 0.712116 | 0.708375 |
| COTAN | 0.716421 | 0.637108 | 0.744518 | 0.690367 | 0.703698 |
print_scores(tuning = 'antibody',dataset="PBMC2")'PBMC2 - number of clusters'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 11 | 10 | 12 | 12 | 12 |
'PBMC2 - Silhuette (higher is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | antibody | |
|---|---|---|---|---|---|---|
| 0 | -0.041184 | 0.037491 | -0.021431 | 0.091472 | 0.074925 | 0.042144 |
'PBMC2 - davies_bouldin (lower is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | antibody | |
|---|---|---|---|---|---|---|
| 0 | 3.22287 | 1.702317 | 4.327977 | 1.463008 | 2.077179 | 2.2928 |
'PBMC2 - matching antibody labels'| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.590346 | 0.458018 | 0.610332 | 0.571627 | 0.544156 |
| scanpy | 0.746517 | 0.649454 | 0.756633 | 0.736667 | 0.707302 |
| scvi-tools | 0.672359 | 0.576869 | 0.708154 | 0.640009 | 0.646073 |
| seurat | 0.759142 | 0.760752 | 0.776572 | 0.742477 | 0.800906 |
| COTAN | 0.735282 | 0.673070 | 0.687591 | 0.790082 | 0.743686 |
print_scores(tuning = 'antibody',dataset="PBMC3")'PBMC3 - number of clusters'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 12 | 14 | 13 | 14 | 12 |
'PBMC3 - Silhuette (higher is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | antibody | |
|---|---|---|---|---|---|---|
| 0 | -0.048747 | 0.024448 | -0.007328 | 0.063501 | 0.05861 | 0.028552 |
'PBMC3 - davies_bouldin (lower is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | antibody | |
|---|---|---|---|---|---|---|
| 0 | 3.082757 | 2.048357 | 2.967632 | 1.617082 | 1.813171 | 2.265731 |
'PBMC3 - matching antibody labels'| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.644484 | 0.537094 | 0.639574 | 0.649469 | 0.598577 |
| scanpy | 0.729603 | 0.683244 | 0.752370 | 0.708173 | 0.724410 |
| scvi-tools | 0.726492 | 0.670063 | 0.728625 | 0.724372 | 0.713239 |
| seurat | 0.764843 | 0.698339 | 0.799673 | 0.732920 | 0.738860 |
| COTAN | 0.691237 | 0.607331 | 0.643954 | 0.746015 | 0.676860 |
print_scores(tuning = 'antibody',dataset="PBMC4")'PBMC4 - number of clusters'| monocle | scanpy | scvi-tools | seurat | COTAN | |
|---|---|---|---|---|---|
| 0 | 13 | 11 | 11 | 13 | 12 |
'PBMC4 - Silhuette (higher is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | antibody | |
|---|---|---|---|---|---|---|
| 0 | -0.009349 | 0.036425 | 0.038929 | 0.059369 | 0.03044 | -0.044213 |
'PBMC4 - davies_bouldin (lower is better)'| monocle | scanpy | scvi-tools | seurat | COTAN | antibody | |
|---|---|---|---|---|---|---|
| 0 | 2.412446 | 1.784603 | 1.670777 | 1.621226 | 1.822676 | 4.131438 |
'PBMC4 - matching antibody labels'| NMI | ARI | homogeneity | completeness | fowlkes_mallows | |
|---|---|---|---|---|---|
| monocle | 0.642736 | 0.462311 | 0.694650 | 0.598042 | 0.558509 |
| scanpy | 0.718317 | 0.585612 | 0.758534 | 0.682150 | 0.662578 |
| scvi-tools | 0.730060 | 0.588797 | 0.763135 | 0.699732 | 0.665395 |
| seurat | 0.722468 | 0.570140 | 0.785446 | 0.668840 | 0.651248 |
| COTAN | 0.675609 | 0.518293 | 0.678118 | 0.673118 | 0.606106 |
Check cellTypist vs Antibody
def compute_clustering_scores(output_dir, dataset):#celltypist_df, antibody_df,
# Merge the dataframes on the common 'cell' column
#cotan_df = pd.read_csv(f'{DIR}{dataset}/COTAN/antibody/clustering_labels.csv', index_col=0)
#display("Cotan clusters objetc dimension ",cotan_df.shape)
#display("----------------------------------------")
celltypist_df = pd.read_csv(f'{DIR}{dataset}/celltypist/celltypist_labels.csv', index_col=0)
celltypist_df.index = celltypist_df.index.str[:-2]
antibody_df = pd.read_csv(f'{DIR}{dataset}/antibody_annotation/antibody_labels_postproc.csv', index_col=0)
#antibody_df = labels_df.merge(antibody_df, how='inner', on='cell')
#all_in_antibody = celltypist_df.index.isin(antibody_df.index).all()
#all_in_celltypist = antibody_df.index.isin(celltypist_df.index).all()
#display("All celltypist indices in antibody: ",all_in_antibody, celltypist_df.index.isin(antibody_df.index).sum(),celltypist_df.shape)
#display("All antibody indices in cellTypist:", all_in_celltypist)
#display("----------------------------------------")
merged_df = celltypist_df.merge(antibody_df, how='inner',left_index=True, right_index=True)# on='cell')
merged_df.columns = ['cluster_celltypist','cluster_antibody']
# Initialize scores dictionary
scores = {
'NMI': normalized_mutual_info_score(merged_df['cluster_celltypist'], merged_df['cluster_antibody'], average_method='arithmetic'),
'ARI': adjusted_rand_score(merged_df['cluster_celltypist'], merged_df['cluster_antibody']),
'Homogeneity': homogeneity_score(merged_df['cluster_celltypist'], merged_df['cluster_antibody']),
'Completeness': completeness_score(merged_df['cluster_celltypist'], merged_df['cluster_antibody']),
'Fowlkes_Mallows': fowlkes_mallows_score(merged_df['cluster_celltypist'], merged_df['cluster_antibody'])
}
# Convert scores to DataFrame
scores_df = pd.DataFrame([scores])
# Save scores to CSV and LaTeX
#scores_df.to_csv(f'{output_dir}{dataset}/clustering_comparison_scores.csv')
#scores_df.to_latex(f'{output_dir}{dataset}/clustering_comparison_scores.tex')
# Display scores DataFrame
display(scores_df)
for dataset in DATASET_NAMES:
#display('------------------------------')
display(f'{dataset} - Clustering Comparison between CellTypist and Antibody')
# Assuming celltypist_df and antibody_df are defined elsewhere and available here
compute_clustering_scores(DIR, dataset)'PBMC1 - Clustering Comparison between CellTypist and Antibody'| NMI | ARI | Homogeneity | Completeness | Fowlkes_Mallows | |
|---|---|---|---|---|---|
| 0 | 0.752326 | 0.731095 | 0.708308 | 0.802178 | 0.78126 |
'PBMC2 - Clustering Comparison between CellTypist and Antibody'| NMI | ARI | Homogeneity | Completeness | Fowlkes_Mallows | |
|---|---|---|---|---|---|
| 0 | 0.659259 | 0.481537 | 0.667725 | 0.651004 | 0.585734 |
'PBMC3 - Clustering Comparison between CellTypist and Antibody'| NMI | ARI | Homogeneity | Completeness | Fowlkes_Mallows | |
|---|---|---|---|---|---|
| 0 | 0.693433 | 0.555502 | 0.693429 | 0.693436 | 0.618105 |
'PBMC4 - Clustering Comparison between CellTypist and Antibody'| NMI | ARI | Homogeneity | Completeness | Fowlkes_Mallows | |
|---|---|---|---|---|---|
| 0 | 0.751294 | 0.7252 | 0.728817 | 0.775201 | 0.776972 |
Summary
External measures
def load_scores(tuning, dataset):
scores = pd.read_csv(f'{DIR}{dataset}/scores_{tuning}.csv')
scores = scores.rename(columns={"Unnamed: 0": "tool"})
scores['tuning'] = tuning
return scoresdatasets = ['PBMC1', 'PBMC2', 'PBMC3', 'PBMC4']
tunings = ['default_celltypist', 'default_antibody', 'celltypist_celltypist', 'antibody_antibody']
scores_list = []
# Concatenate all scores into one DataFrame
for dataset in datasets:
for tuning in tunings:
scores = load_scores(tuning, dataset)
scores['dataset'] = dataset
scores_list.append(scores)
all_scores = pd.concat(scores_list)
# Prepare data for plotting
all_scores_melted = all_scores.melt(id_vars=['tool', 'tuning', 'dataset'], var_name='score', value_name='value')
sns.set_context("talk")
# Define custom colors
custom_palette = {
"seurat": "#4575B4",
"monocle": "#DAABE9",
"scanpy": "#7F9B5C",
"COTAN": "#F73604",
"scvi-tools": "#B6A18F"
}
g = sns.FacetGrid(all_scores_melted, row='score', col='tuning', sharey=False, height=4, aspect=1.3)
g.map(sns.pointplot, 'tool', 'value', palette=custom_palette,capsize=0.2, errwidth=2)
# Set titles and labels
g.set_titles(col_template="{col_name}", row_template="{row_name}")
g.set_axis_labels("Tool", "Score Value")
plt.subplots_adjust(top=1.4)
#g.fig.suptitle('Comparison of Clustering Tools by Various Scores and Conditions')
# Rotate x-axis labels
for ax in g.axes.flatten():
plt.setp(ax.get_xticklabels(), rotation=45)
g.savefig("ClusteringToolsComparison.pdf")
plt.show()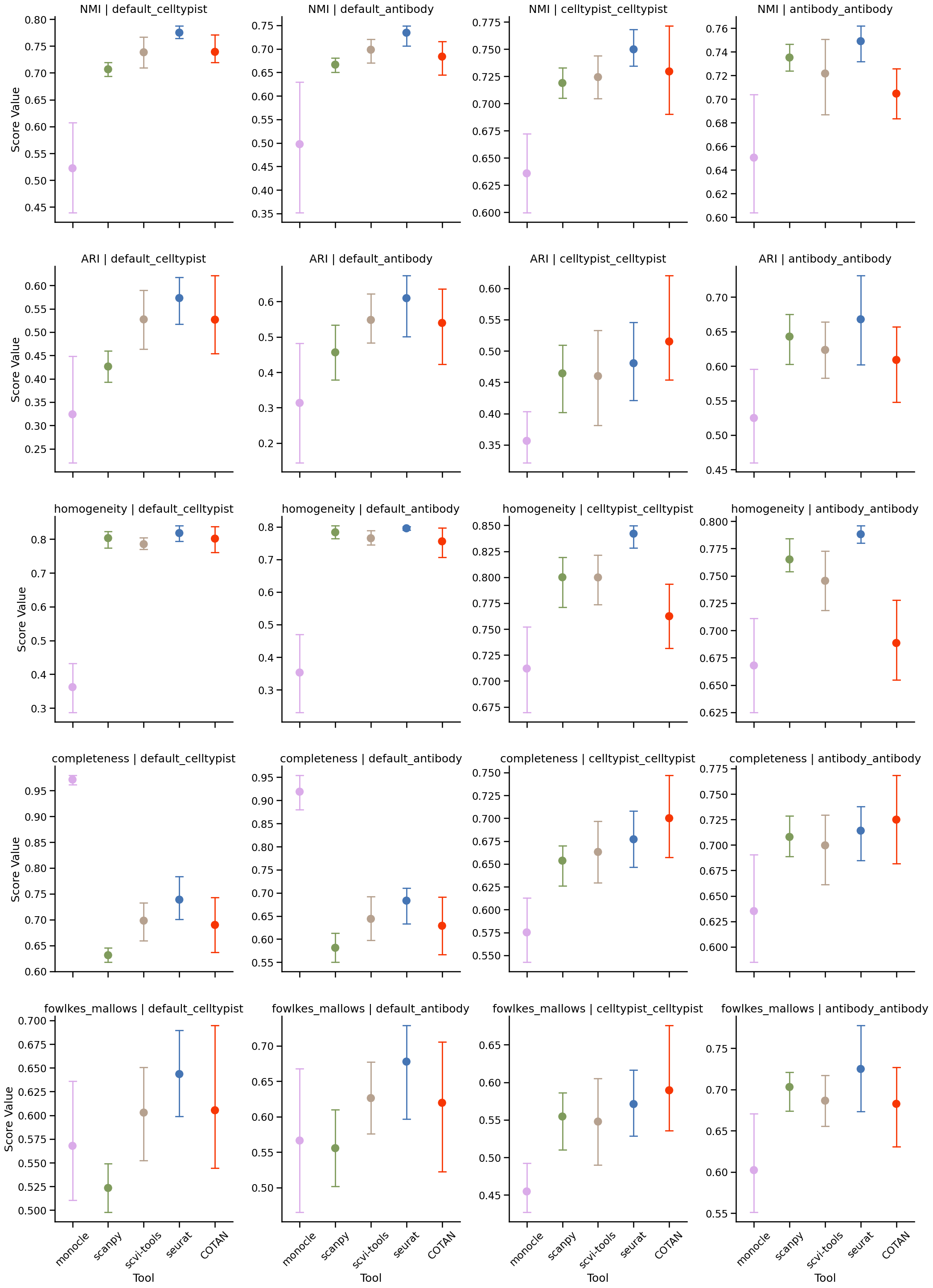
Internal measures
# Load your data (assuming you have CSV files for the scores)
def load_scores(tuning, dataset, score_type):
file_path = f'{DIR}{dataset}/{tuning}_{score_type}.csv'
print(f"Loading {file_path}")
scores = pd.read_csv(file_path, header=0) # Read the CSV file without an index column
scores_melted = scores.melt(var_name='tool', value_name='value')
scores_melted['tuning'] = tuning
scores_melted['dataset'] = dataset
scores_melted['score_type'] = score_type
return scores_melted
datasets = ['PBMC1', 'PBMC2', 'PBMC3', 'PBMC4']
tunings = ['default', 'celltypist', 'antibody']
score_types = ['silhouette', 'davies_bouldin']
scores_list = []
# Concatenate all scores into one DataFrame
for dataset in datasets:
for tuning in tunings:
for score_type in score_types:
scores = load_scores(tuning, dataset, score_type)
scores_list.append(scores)
all_scores = pd.concat(scores_list)
# Debug: Check the loaded data
print(all_scores.head())
# Define custom colors
custom_palette = {
"seurat": "#4575B4",
"monocle": "#DAABE9",
"scanpy": "#7F9B5C",
"COTAN": "#F73604",
"scvi-tools": "#B6A18F"
}
# Filter for silhouette and davies_bouldin scores
silhouette_scores = all_scores[all_scores['score_type'] == 'silhouette']
davies_bouldin_scores = all_scores[all_scores['score_type'] == 'davies_bouldin']
# Plot Silhouette scores
g1 = sns.FacetGrid(silhouette_scores, col='tuning', sharey=False, height=4, aspect=1.8)
g1.map(sns.pointplot, 'tool', 'value', palette=custom_palette, order=[ "monocle", "scanpy", "scvi-tools","seurat","COTAN"],capsize=0.2, errwidth=2)
g1.set_titles(col_template="{col_name}")
g1.set_axis_labels("Tool", "Silhouette Score")
g1.fig.suptitle('Silhouette Scores by Tool and Tuning Condition', y=1.25)
plt.subplots_adjust(top=0.85)
# Rotate x-axis labels
for ax in g1.axes.flatten():
plt.setp(ax.get_xticklabels(), rotation=45)
# Plot Davies-Bouldin scores
g2 = sns.FacetGrid(davies_bouldin_scores, col='tuning', sharey=False, height=4, aspect=1.8)
g2.map(sns.pointplot, 'tool', 'value', palette=custom_palette, order=["monocle", "scanpy", "scvi-tools","seurat","COTAN"],capsize=0.2, errwidth=2)
g2.set_titles(col_template="{col_name}")
g2.set_axis_labels("Tool", "Davies-Bouldin Score")
g2.fig.suptitle('Davies-Bouldin Scores by Tool and Tuning Condition', y=1.85)
plt.subplots_adjust(top=1.5)
# Rotate x-axis labels
for ax in g2.axes.flatten():
plt.setp(ax.get_xticklabels(), rotation=45)
g1.savefig("ClusteringToolsSilhouette.png")
g2.savefig("ClusteringToolsDaviesBouldin.png")
plt.show()Loading Data/PBMC1/default_silhouette.csv
Loading Data/PBMC1/default_davies_bouldin.csv
Loading Data/PBMC1/celltypist_silhouette.csv
Loading Data/PBMC1/celltypist_davies_bouldin.csv
Loading Data/PBMC1/antibody_silhouette.csv
Loading Data/PBMC1/antibody_davies_bouldin.csv
Loading Data/PBMC2/default_silhouette.csv
Loading Data/PBMC2/default_davies_bouldin.csv
Loading Data/PBMC2/celltypist_silhouette.csv
Loading Data/PBMC2/celltypist_davies_bouldin.csv
Loading Data/PBMC2/antibody_silhouette.csv
Loading Data/PBMC2/antibody_davies_bouldin.csv
Loading Data/PBMC3/default_silhouette.csv
Loading Data/PBMC3/default_davies_bouldin.csv
Loading Data/PBMC3/celltypist_silhouette.csv
Loading Data/PBMC3/celltypist_davies_bouldin.csv
Loading Data/PBMC3/antibody_silhouette.csv
Loading Data/PBMC3/antibody_davies_bouldin.csv
Loading Data/PBMC4/default_silhouette.csv
Loading Data/PBMC4/default_davies_bouldin.csv
Loading Data/PBMC4/celltypist_silhouette.csv
Loading Data/PBMC4/celltypist_davies_bouldin.csv
Loading Data/PBMC4/antibody_silhouette.csv
Loading Data/PBMC4/antibody_davies_bouldin.csv
tool value tuning dataset score_type
0 Unnamed: 0 0.000000 default PBMC1 silhouette
1 monocle 0.100032 default PBMC1 silhouette
2 scanpy 0.043761 default PBMC1 silhouette
3 scvi-tools 0.065534 default PBMC1 silhouette
4 seurat 0.148254 default PBMC1 silhouette