library(COTAN)
library(ComplexHeatmap)
library(circlize)
library(dplyr)
library(Hmisc)
library(Seurat)
library(patchwork)
library(Rfast)
library(parallel)
library(doParallel)
library(HiClimR)
library(stringr)
library(fst)
options(parallelly.fork.enable = TRUE)
dataFile <- "Data/Brown_PBMC_Datasets/capillary_blood_samples_pbmcs-Run_40-Cleaned.RDS"
name <- str_split(dataFile,pattern = "/",simplify = T)[3]
name <- str_remove(name,pattern = "-Cleaned.RDS")
name <- str_remove(name,pattern = "capillary_blood_samples_")
project = name
setLoggingLevel(1)
outDir <- "CoexData/"
setLoggingFile(paste0(outDir, "Logs/",name,".log"))
obj <- readRDS(dataFile)
file_code = nameGene Correlation Analysis for PBMC Brown run 40
Prologue
source("src/Functions.R")To compare the ability of COTAN to asses the real correlation between genes we define some pools of genes:
- Constitutive genes
- Neural progenitor genes
- Pan neuronal genes
- Some layer marker genes
hkGenes <- read.csv("Data/Housekeeping_TranscriptsHuman.csv", sep = ";")
genesList <- list(
"CD4 T cells"=
c("TRBC2","CD3D","CD3G","CD3E","IL7R","LTB","LEF1"),
"B cells"=
c("PAX5","MS4A1","CD19","CD74","CD79A","IGHD","HLA-DRA"),
"hk"= hkGenes$Gene_symbol[1:20], # from https://housekeeping.unicamp.br/
"general PBMC markers" =
c("FOXP3","TBX21","GATA3","RUNX1","BCL6","EOMES","EOMES","TBX21","BATF3","IRF2","TCF4","STAT5A","RUNX3","STAT6","BATF","STAT3","TBX21","TBX21","IRF8","IRF4","AHR","STAT1","IRF4","RELB")
)COTAN
genesFromListExpressed <- unlist(genesList)[unlist(genesList) %in% getGenes(obj)]
int.genes <-getGenes(obj)obj <- proceedToCoex(obj, calcCoex = TRUE, cores = 5L, saveObj = FALSE)coexMat.big <- getGenesCoex(obj)[genesFromListExpressed,genesFromListExpressed]
coexMat <- getGenesCoex(obj)[c(genesList$`CD4 T cells`,genesList$hk,genesList$`B cells`),c(genesList$`CD4 T cells`,genesList$hk,genesList$`B cells`)]
f1 = colorRamp2(seq(-1,1, length = 3), c("#DC0000B2", "white","#3C5488B2" ))
split.genes <- base::factor(c(rep("CD4 T cells",length(genesList[["CD4 T cells"]])),
rep("HK",length(genesList[["hk"]])),
rep("B cells",length(genesList[["B cells"]]))
),
levels = c("CD4 T cells","HK","B cells"))
lgd = Legend(col_fun = f1, title = "COTAN coex")
htmp <- Heatmap(as.matrix(coexMat),
#width = ncol(coexMat)*unit(2.5, "mm"),
height = nrow(coexMat)*unit(3, "mm"),
cluster_rows = FALSE,
cluster_columns = FALSE,
col = f1,
row_names_side = "left",
row_names_gp = gpar(fontsize = 11),
column_names_gp = gpar(fontsize = 11),
column_split = split.genes,
row_split = split.genes,
cluster_row_slices = FALSE,
cluster_column_slices = FALSE,
heatmap_legend_param = list(
title = "COTAN coex", at = c(-1, 0, 1),
direction = "horizontal",
labels = c("-1", "0", "1")
)
)
draw(htmp, heatmap_legend_side="right")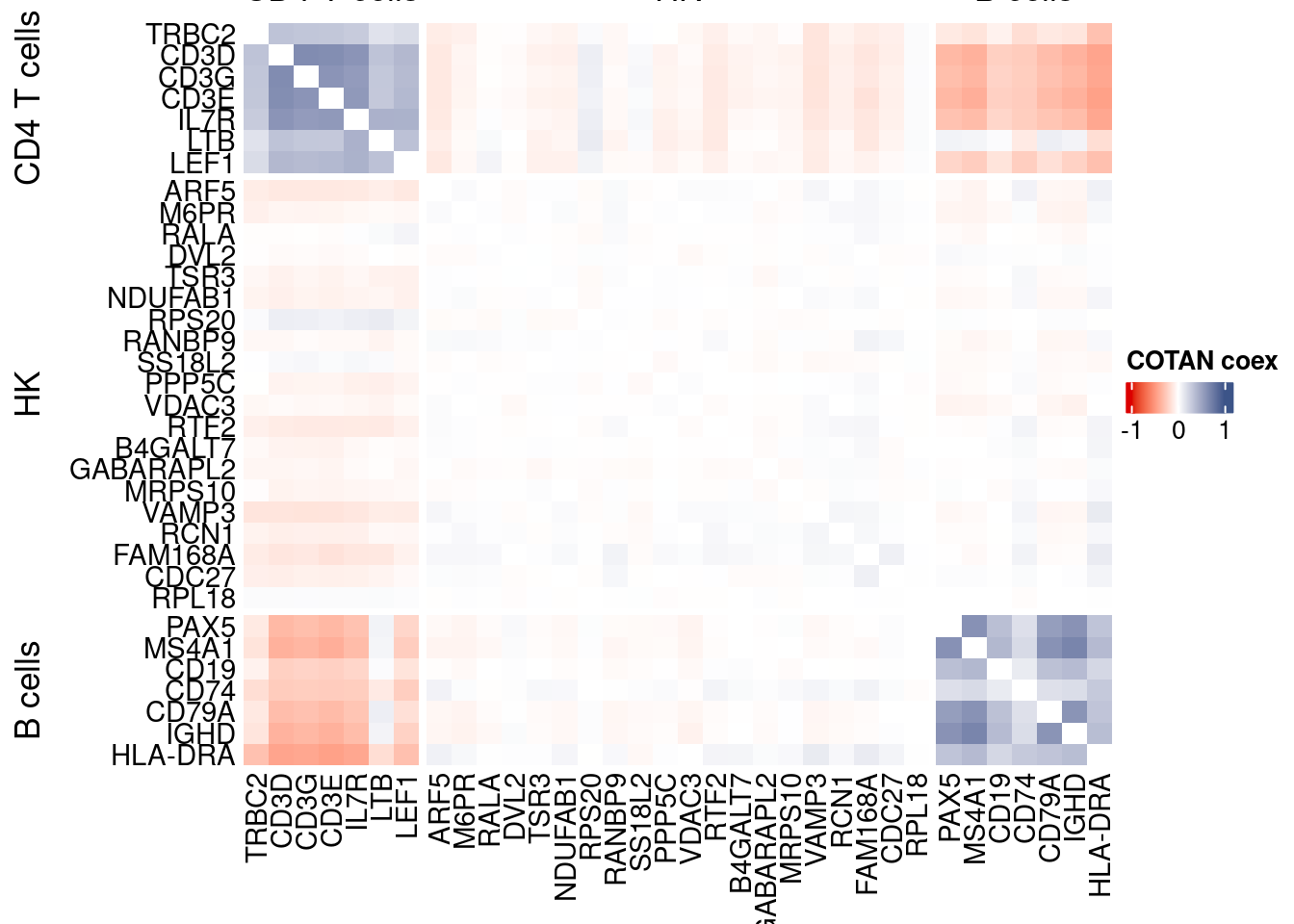
GDI_DF <- calculateGDI(obj)
GDI_DF$geneType <- NA
for (cat in names(genesList)) {
GDI_DF[rownames(GDI_DF) %in% genesList[[cat]],]$geneType <- cat
}
GDI_DF$GDI_centered <- scale(GDI_DF$GDI,center = T,scale = T)
GDI_DF[genesFromListExpressed,] sum.raw.norm GDI exp.cells geneType GDI_centered
TRBC2 10.281423 4.6753458 79.580831 CD4 T cells 2.38465433
CD3D 10.138257 5.1870513 79.568575 CD4 T cells 2.88961161
CD3G 9.947640 5.1005711 76.443192 CD4 T cells 2.80427181
CD3E 10.028351 5.1840570 79.740164 CD4 T cells 2.88665672
IL7R 10.819269 5.0363720 75.903910 CD4 T cells 2.74091943
LTB 11.066033 4.7167109 86.138007 CD4 T cells 2.42547392
LEF1 9.575518 4.5923187 55.742125 CD4 T cells 2.30272214
PAX5 6.594448 3.5008847 3.909793 B cells 1.22568178
MS4A1 7.982075 3.6449369 7.022919 B cells 1.36783428
CD19 5.271515 2.7354892 1.703640 B cells 0.47038014
CD74 10.236047 3.9041546 50.667974 B cells 1.62363341
CD79A 8.405070 3.5013672 9.094252 B cells 1.22615792
IGHD 7.579516 3.5736365 4.694203 B cells 1.29747416
HLA-DRA 9.889789 4.9280298 22.147322 B cells 2.63400602
ARF5 8.182166 2.9943936 34.526290 hk 0.72587022
M6PR 8.465049 2.4504986 42.578747 hk 0.18914797
RALA 7.982402 2.0919776 28.741267 hk -0.16464490
DVL2 5.942692 1.3727948 4.252972 hk -0.87434332
TSR3 7.545354 1.7245626 20.321118 hk -0.52721453
NDUFAB1 8.375120 2.3293029 40.973158 hk 0.06955059
RPS20 11.476512 2.0116638 99.497487 hk -0.24389958
RANBP9 7.930775 2.8666734 26.302243 hk 0.59983437
SS18L2 8.243229 1.9083038 36.524084 hk -0.34589648
PPP5C 6.508700 1.8772139 7.353842 hk -0.37657639
VDAC3 8.144980 1.6578412 33.410957 hk -0.59305603
RTF2 8.117900 2.6697077 33.374188 hk 0.40546618
B4GALT7 6.120160 1.9704804 5.270254 hk -0.28453982
GABARAPL2 8.674787 1.9170136 50.667974 hk -0.33730155
MRPS10 7.511741 1.4972229 19.107734 hk -0.75155618
VAMP3 6.885916 3.4147202 11.288148 hk 1.14065363
RCN1 6.014452 2.3955685 4.951587 hk 0.13494228
FAM168A 7.348677 3.9478265 14.156147 hk 1.66672944
CDC27 7.735180 3.0692921 22.784655 hk 0.79978093
RPL18 12.097345 0.8765238 99.975487 hk -1.36406958
FOXP3 4.997158 1.7170950 1.299179 general PBMC markers -0.53458364
TBX21 6.922577 3.3540603 7.451894 general PBMC markers 1.08079369
GATA3 7.804577 2.8024667 21.559015 general PBMC markers 0.53647440
RUNX1 8.887512 3.9358183 47.898027 general PBMC markers 1.65487958
BCL6 6.501857 4.4578044 7.059689 general PBMC markers 2.16998179
EOMES 6.364606 2.7428021 5.061895 general PBMC markers 0.47759662
EOMES.1 6.364606 2.7428021 5.061895 general PBMC markers 0.47759662
TBX21.1 6.922577 3.3540603 7.451894 general PBMC markers 1.08079369
BATF3 4.644769 3.8548176 1.397230 general PBMC markers 1.57494705
IRF2 8.418316 2.2969735 40.850594 general PBMC markers 0.03764747
TCF4 6.725206 3.9874305 5.282510 general PBMC markers 1.70581113
STAT5A 6.907744 2.4855603 11.459738 general PBMC markers 0.22374732
RUNX3 8.126983 2.7185042 27.442088 general PBMC markers 0.45361918
STAT6 7.391468 3.7597434 17.526658 general PBMC markers 1.48112672
BATF 7.519322 2.2810830 17.428606 general PBMC markers 0.02196662
STAT3 8.284076 3.1100404 36.033828 general PBMC markers 0.83999188
TBX21.2 6.922577 3.3540603 7.451894 general PBMC markers 1.08079369
TBX21.3 6.922577 3.3540603 7.451894 general PBMC markers 1.08079369
IRF8 6.837432 4.5620196 6.949381 general PBMC markers 2.27282265
IRF4 5.242881 3.1579831 2.157127 general PBMC markers 0.88730232
AHR 7.442754 4.6313180 13.334968 general PBMC markers 2.34120721
STAT1 8.519977 2.8188197 33.839931 general PBMC markers 0.55261173
IRF4.1 5.242881 3.1579831 2.157127 general PBMC markers 0.88730232
RELB 6.831197 3.7263740 10.172815 general PBMC markers 1.44819742GDIPlot(obj,GDIIn = GDI_DF, genes = genesList,GDIThreshold = 1.4)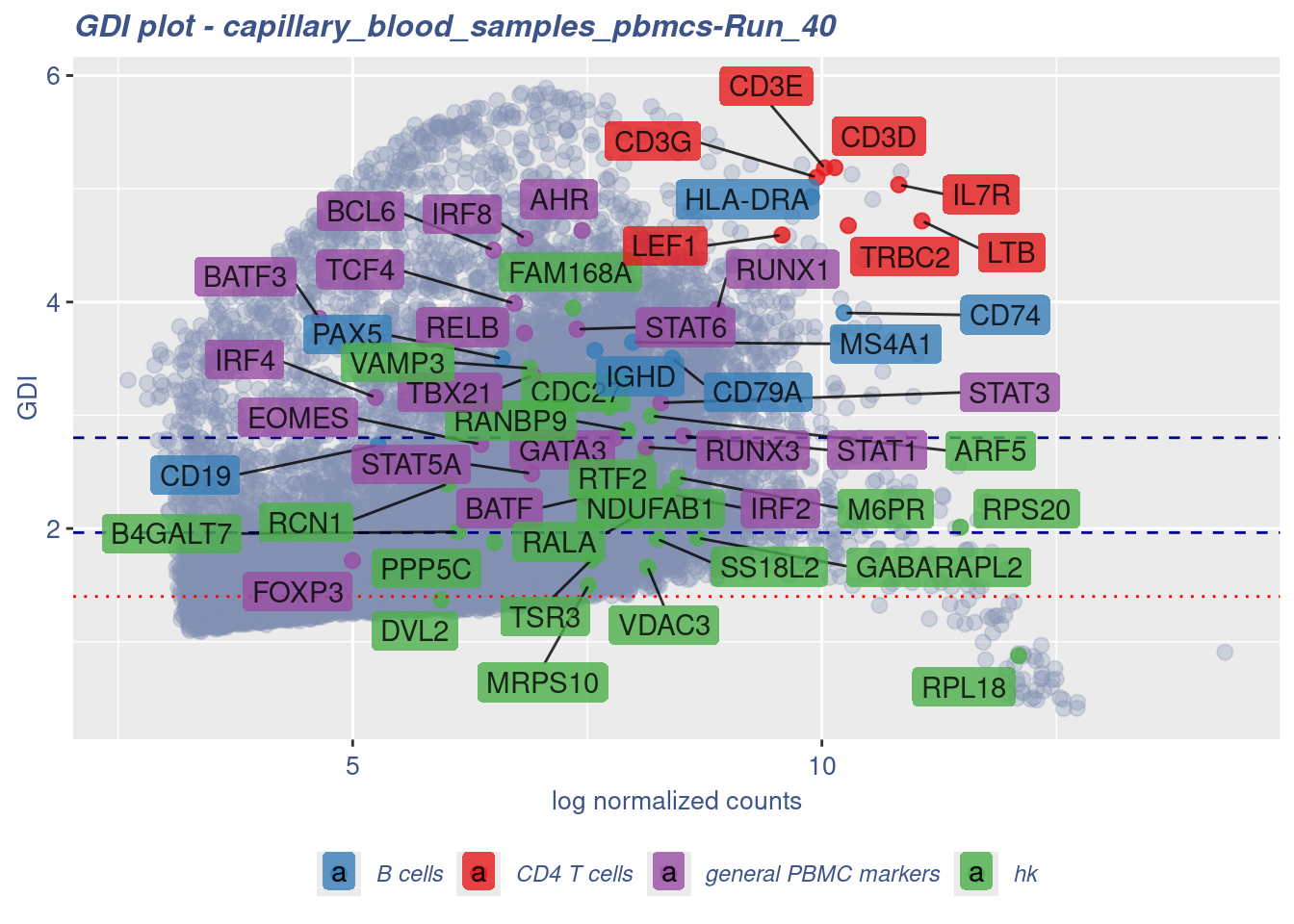
Seurat correlation
srat<- CreateSeuratObject(counts = getRawData(obj),
project = project,
min.cells = 3,
min.features = 200)
srat[["percent.mt"]] <- PercentageFeatureSet(srat, pattern = "^MT-")
srat <- NormalizeData(srat)
srat <- FindVariableFeatures(srat, selection.method = "vst", nfeatures = 2000)
# plot variable features with and without labels
plot1 <- VariableFeaturePlot(srat)
plot1$data$centered_variance <- scale(plot1$data$variance.standardized,
center = T,scale = F)
write.csv(plot1$data,paste0("CoexData/",
"Variance_Seurat_genes",
getMetadataElement(obj,file_code),".csv"))
LabelPoints(plot = plot1, points = c(genesList$`CD4 T cells`,genesList$`B cells`,genesList$hk), repel = TRUE)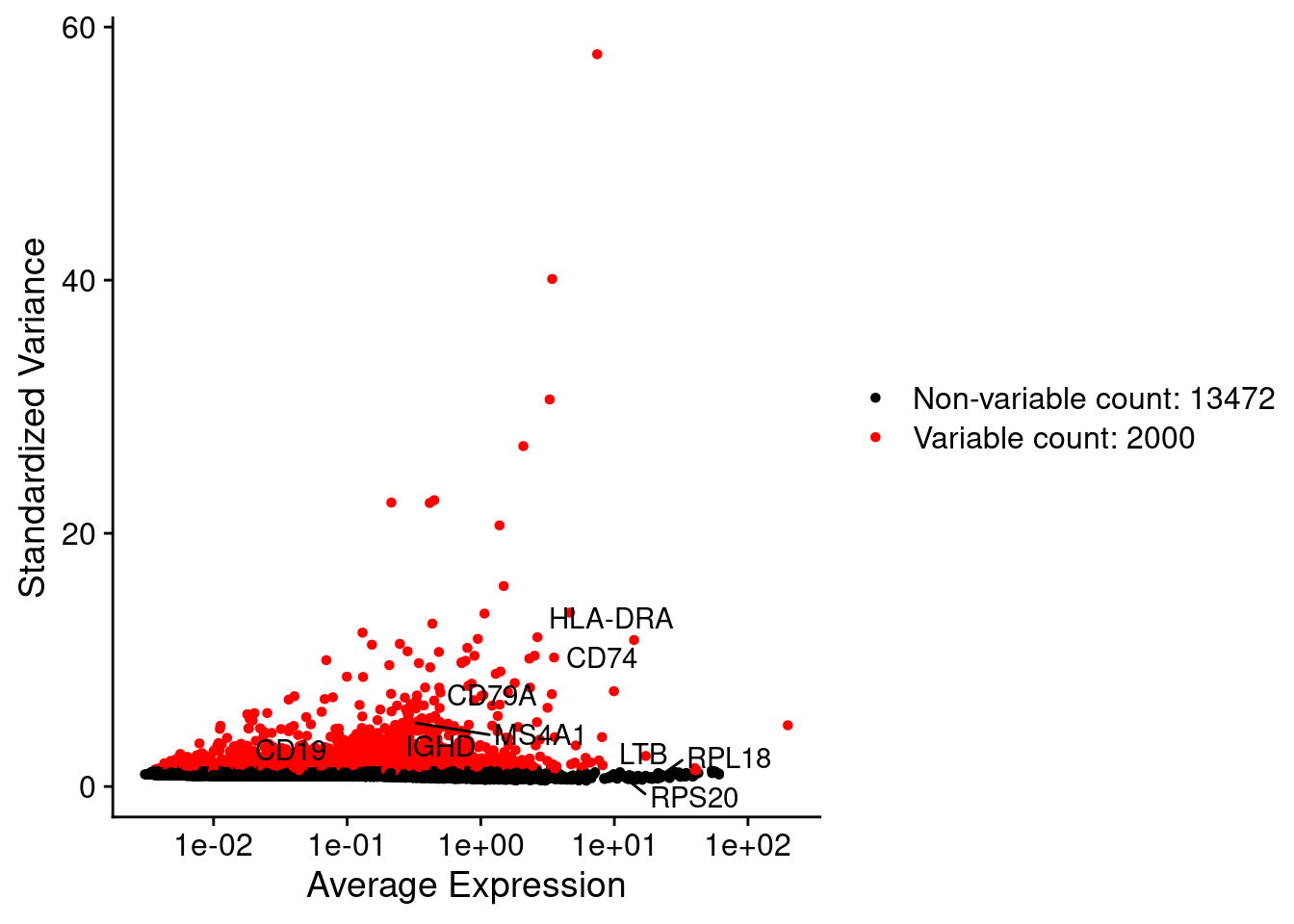
LabelPoints(plot = plot1, points = c(genesList$hk), repel = TRUE)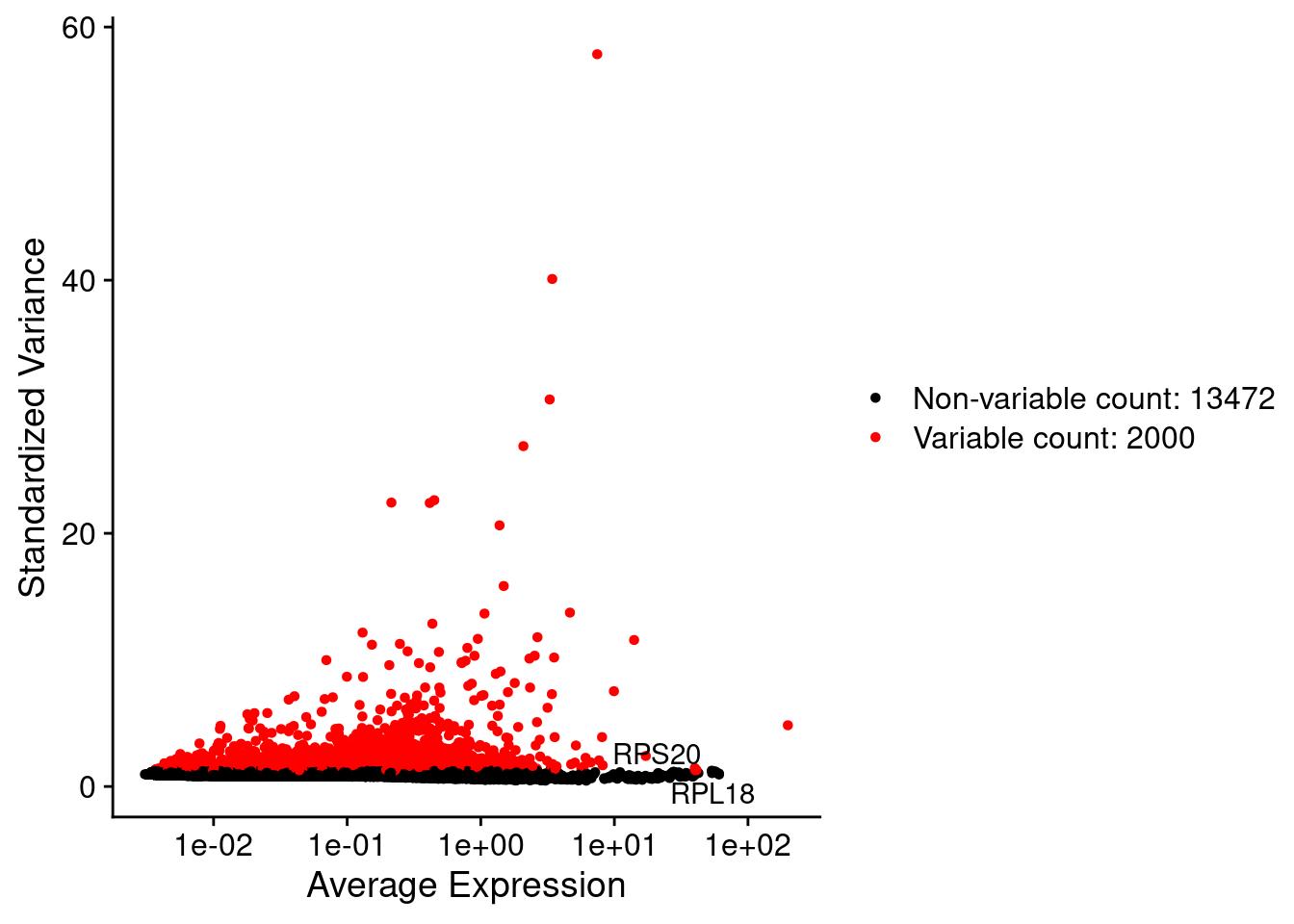
all.genes <- rownames(srat)
srat <- ScaleData(srat, features = all.genes)
seurat.data = GetAssayData(srat[["RNA"]],layer = "data")corr.pval.list <- correlation_pvalues(data = seurat.data,
genesFromListExpressed,
n.cells = getNumCells(obj))
seurat.data.cor.big <- as.matrix(Matrix::forceSymmetric(corr.pval.list$data.cor, uplo = "U"))
htmp <- correlation_plotPBMC(seurat.data.cor.big,
genesList, title="Seurat corr")
p_values.fromSeurat <- corr.pval.list$p_values
seurat.data.cor.big <- corr.pval.list$data.cor
rm(corr.pval.list)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 10572443 564.7 17840978 952.9 17840978 952.9
Vcells 360781135 2752.6 731877821 5583.8 1478343794 11278.9draw(htmp, heatmap_legend_side="right")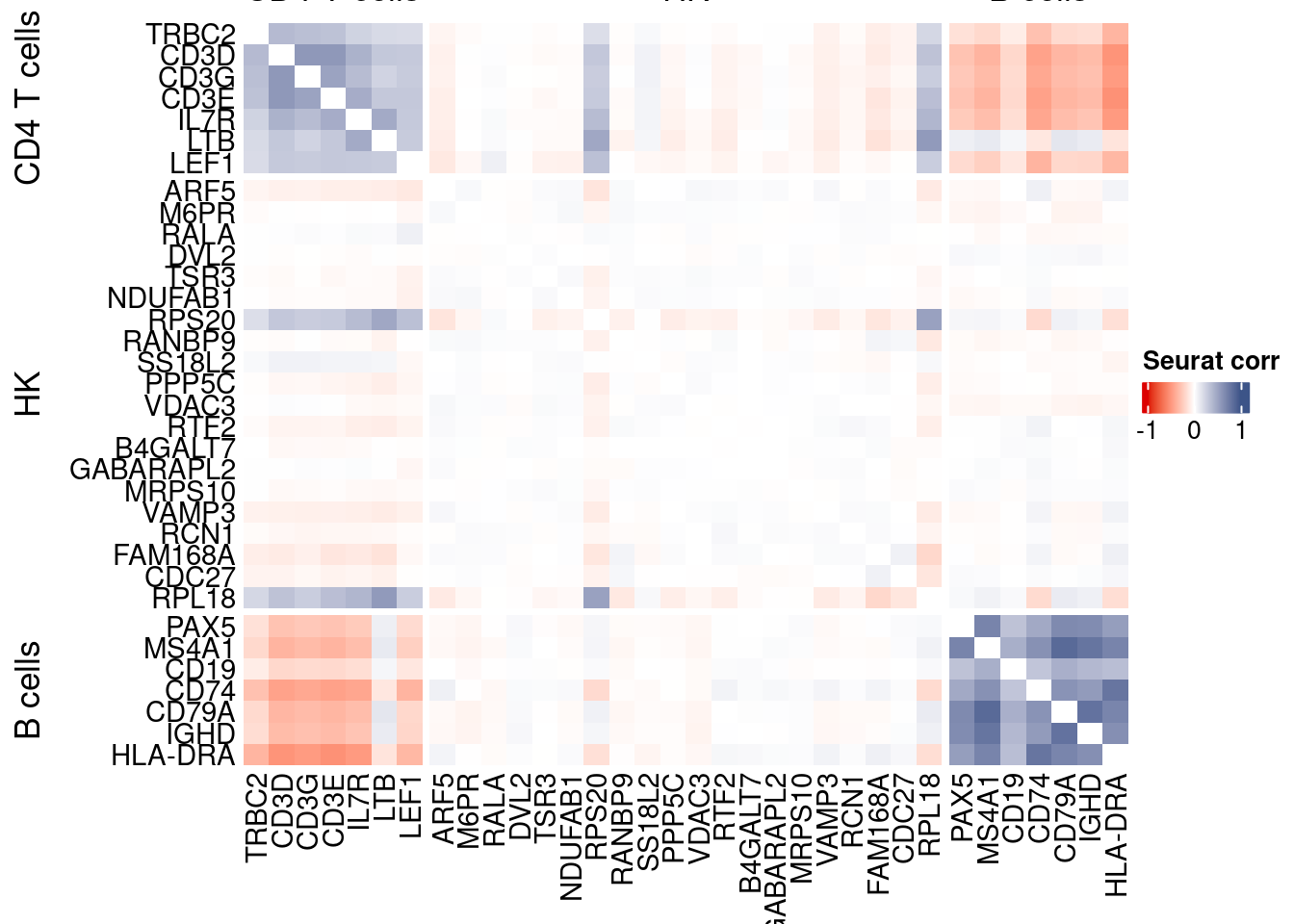
rm(seurat.data.cor.big)
rm(p_values.fromSeurat)
sratbase <- sratSeurat SC Transform
srat <- SCTransform(srat,
method = "glmGamPoi",
vars.to.regress = "percent.mt",
verbose = FALSE)
seurat.data <- GetAssayData(srat[["SCT"]],layer = "data")
#Remove genes with all zeros
seurat.data <-seurat.data[rowSums(seurat.data) > 0,]
corr.pval.list <- correlation_pvalues(seurat.data,
genesFromListExpressed,
n.cells = getNumCells(obj))
seurat.data.cor.big <- as.matrix(Matrix::forceSymmetric(corr.pval.list$data.cor, uplo = "U"))
htmp <- correlation_plotPBMC(seurat.data.cor.big,
genesList, title="Seurat corr SCT")
p_values.fromSeurat <- corr.pval.list$p_values
seurat.data.cor.big <- corr.pval.list$data.cor
rm(corr.pval.list)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 10900656 582.2 17840978 952.9 17840978 952.9
Vcells 432266058 3298.0 1054080062 8042.0 1478343794 11278.9draw(htmp, heatmap_legend_side="right")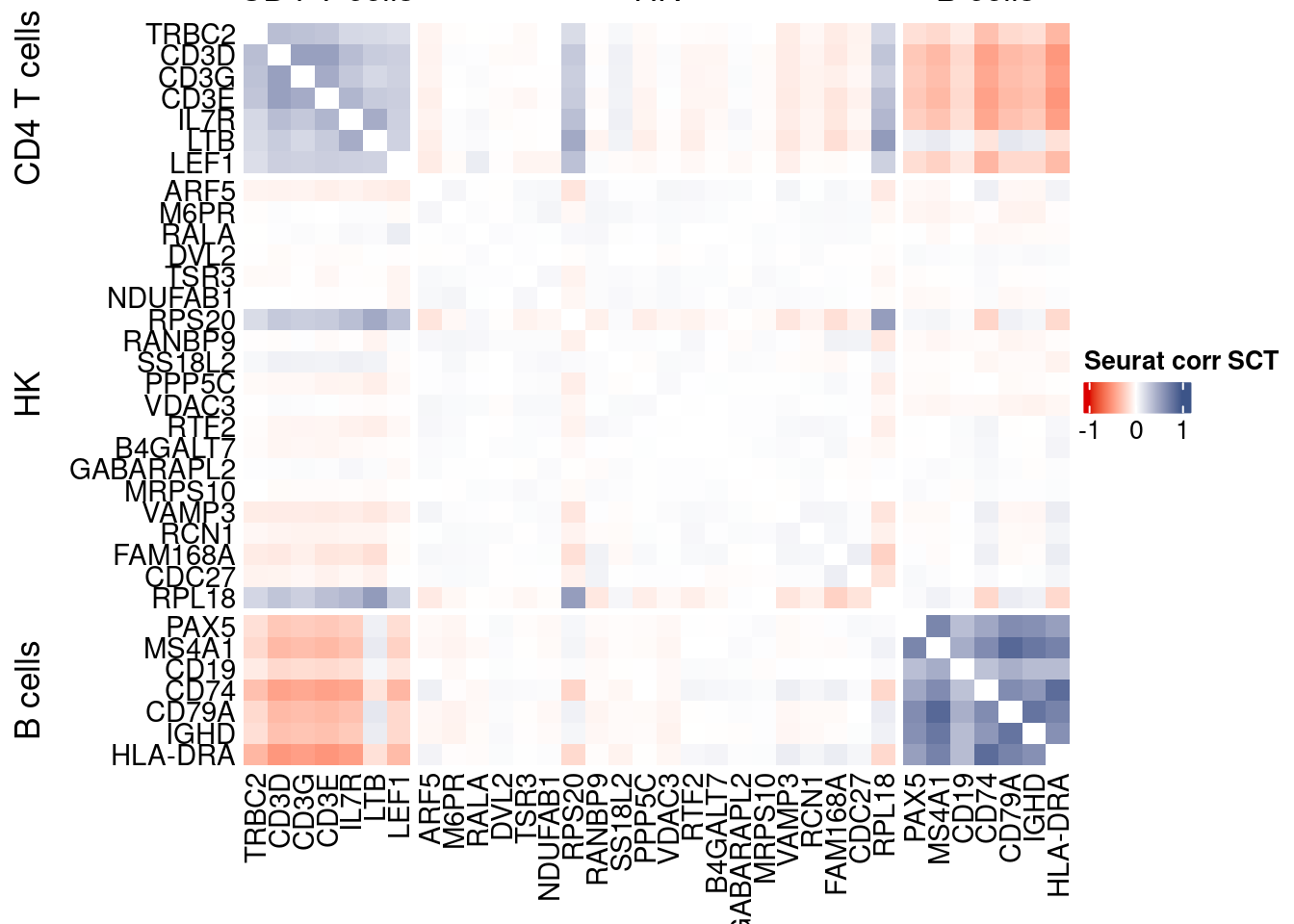
plot1 <- VariableFeaturePlot(srat)
plot1$data$centered_variance <- scale(plot1$data$residual_variance,
center = T,scale = F)write.csv(plot1$data,paste0("CoexData/",
"Variance_SeuratSCT_genes",
file_code,".csv"))
write_fst(as.data.frame(seurat.data.cor.big),path = paste0("CoexData/SeuratCorrSCT_",file_code,".fst"), compress = 100)
write_fst(as.data.frame(p_values.fromSeurat),path = paste0("CoexData/SeuratPValuesSCT_", file_code,".fst"))
write.csv(as.data.frame(p_values.fromSeurat),paste0("CoexData/SeuratPValuesSCT_", file_code,".csv"))
rm(seurat.data.cor.big)
rm(p_values.fromSeurat)Monocle
library(monocle3)cds <- new_cell_data_set(getRawData(obj),
#cell_metadata = getMetadataCells(obj),
#gene_metadata = getMetadataGenes(obj)
)
cds <- preprocess_cds(cds, num_dim = 100)
normalized_counts <- normalized_counts(cds)#Remove genes with all zeros
normalized_counts <- normalized_counts[rowSums(normalized_counts) > 0,]
corr.pval.list <- correlation_pvalues(normalized_counts,
genesFromListExpressed,
n.cells = getNumCells(obj))
rm(normalized_counts)
monocle.data.cor.big <- as.matrix(Matrix::forceSymmetric(corr.pval.list$data.cor, uplo = "U"))
htmp <- correlation_plotPBMC(data.cor.big = monocle.data.cor.big,
genesList,
title = "Monocle corr")
p_values.from.monocle <- corr.pval.list$p_values
monocle.data.cor.big <- corr.pval.list$data.cor
rm(corr.pval.list)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 11086775 592.1 17840978 952.9 17840978 952.9
Vcells 434993007 3318.8 1054080062 8042.0 1478343794 11278.9draw(htmp, heatmap_legend_side="right")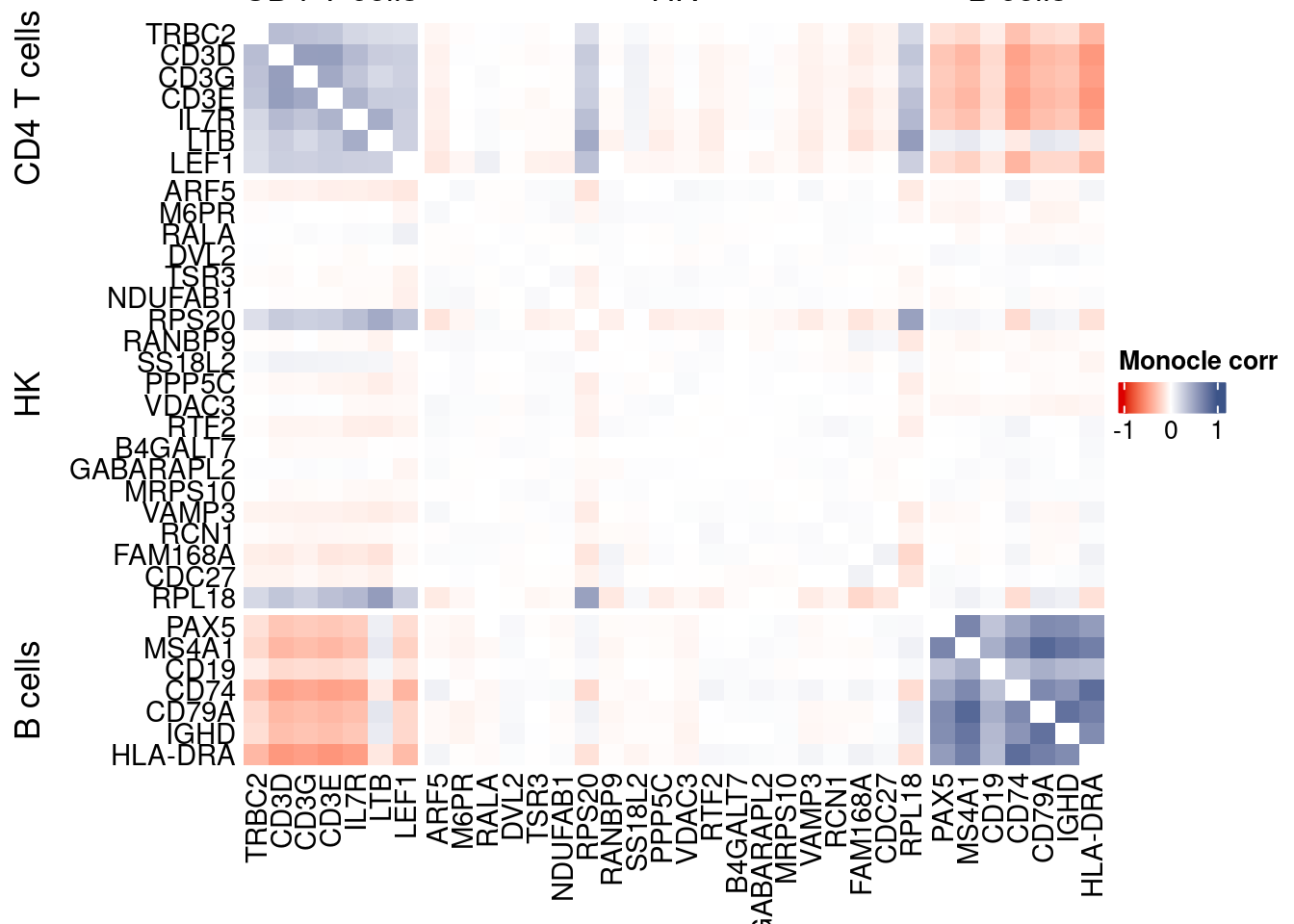
Cs-Core
devtools::load_all("../CS-CORE/")Exports from /mnt/d/CS-CORE/src/CSCORE_IRLS_cpp.cpp:
Rcpp::List CSCORE_IRLS_cpp_impl(const arma::mat& X, const arma::vec& seq_depth_sq, const arma::mat& D_mu, const arma::mat& D_sigma2, const arma::mat& D_sigma, const bool post_process = true, const int n_iter = 10, const double eps = 0.05, const bool verbose = false, const std::string& conv = "q95", const bool return_all = false)
Exports from /mnt/d/CS-CORE/src/WLS_cov.cpp:
Rcpp::List WLS_cov(arma::mat D, arma::mat X, arma::mat W)
Exports from /mnt/d/CS-CORE/src/WLS_mean.cpp:
arma::mat WLS_mean(arma::mat D, arma::mat X, arma::mat W)
/mnt/d/CS-CORE/src/RcppExports.cpp updated.
/mnt/d/CS-CORE/R/RcppExports.R updated.── R CMD INSTALL ───────────────────────────────────────────────────────────────
* installing *source* package ‘CSCORE’ ...
** this is package ‘CSCORE’ version ‘1.0.2’
** using staged installation
** libs
using C++ compiler: ‘g++ (Ubuntu 11.4.0-1ubuntu1~22.04.2) 11.4.0’
g++ -std=gnu++17 -I"/usr/share/R/include" -DNDEBUG -I"/home/seriph/R/x86_64-pc-linux-gnu-library/4.5/RcppArmadillo/include" -I'/home/seriph/R/x86_64-pc-linux-gnu-library/4.5/Rcpp/include' -I'/home/seriph/R/x86_64-pc-linux-gnu-library/4.5/RcppArmadillo/include' -fpic -g -O2 -ffile-prefix-map=/build/r-base-xupQTd/r-base-4.5.2=. -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 -UNDEBUG -Wall -pedantic -g -O0 -c RcppExports.cpp -o RcppExports.o
Error: object 'LdFlags' not found
Execution halted
Error: object 'LdFlags' not found
Execution halted
g++ -std=gnu++17 -shared -L/usr/lib/R/lib -Wl,-Bsymbolic-functions -flto=auto -ffat-lto-objects -flto=auto -Wl,-z,relro -o CSCORE.so CSCORE_IRLS_cpp.o RcppExports.o WLS_cov.o WLS_mean.o -llapack -lblas -lgfortran -lm -lquadmath -L/usr/lib/R/lib -lR
installing to /tmp/RtmpwbZI17/devtools_install_720875f0aef0/00LOCK-CS-CORE/00new/CSCORE/libs
** checking absolute paths in shared objects and dynamic libraries
* DONE (CSCORE)Convert to Seurat obj
sceObj <- convertToSingleCellExperiment(obj)
# Correct: assay=NULL (or omit), data=NULL (since no logcounts)
seuratObj <- as.Seurat(
x = sceObj,
counts = "counts",
data = NULL,
assay = NULL, # IMPORTANT: do NOT set to "RNA" here
project = "COTAN"
)
# as.Seurat(SCE) creates assay "originalexp" by default; rename it to RNA
seuratObj <- RenameAssays(seuratObj, originalexp = "RNA", verbose = FALSE)
DefaultAssay(seuratObj) <- "RNA"
# Optional: keep COTAN payload
seuratObj@misc$COTAN <- S4Vectors::metadata(sceObj)Extract CS_CORE corr matrix
#seuratObj@assays$RNA@counts <- ceiling(seuratObj@assays$RNA@counts)
csCoreRes <- CSCORE(sratbase, genes = genesFromListExpressed)[INFO] IRLS converged after 3 iterations.
[INFO] Starting WLS for covariance at Thu Jan 22 12:35:45 2026
[INFO] 5 among 58 genes have invalid variance estimates. Their co-expressions with other genes were set to 0.
[INFO] 0.7864% co-expression estimates were greater than 1 and were set to 1.
[INFO] 0.0605% co-expression estimates were smaller than -1 and were set to -1.
[INFO] Finished WLS. Elapsed time: 0.8568 seconds.mat <- as.matrix(csCoreRes$est)
diag(mat) <- 0
split.genes <- base::factor(c(rep("CD4 T cells",sum(genesList[["CD4 T cells"]] %in% genesFromListExpressed)),
rep("HK",sum(genesList[["hk"]] %in% genesFromListExpressed)),
rep("B cells",sum(genesList[["B cells"]] %in% genesFromListExpressed))
),
levels = c("CD4 T cells","HK","B cells"))
f1 = colorRamp2(seq(-1,1, length = 3), c("#DC0000B2", "white","#3C5488B2" ))
htmp <- Heatmap(as.matrix(mat[c(genesList$`CD4 T cells`,genesList$hk,genesList$`B cells`),c(genesList$`CD4 T cells`,genesList$hk,genesList$`B cells`)]),
#width = ncol(coexMat)*unit(2.5, "mm"),
height = nrow(mat)*unit(3, "mm"),
cluster_rows = FALSE,
cluster_columns = FALSE,
col = f1,
row_names_side = "left",
row_names_gp = gpar(fontsize = 11),
column_names_gp = gpar(fontsize = 11),
column_split = split.genes,
row_split = split.genes,
cluster_row_slices = FALSE,
cluster_column_slices = FALSE,
heatmap_legend_param = list(
title = "CS-CORE", at = c(-1, 0, 1),
direction = "horizontal",
labels = c("-1", "0", "1")
)
)
draw(htmp, heatmap_legend_side="right")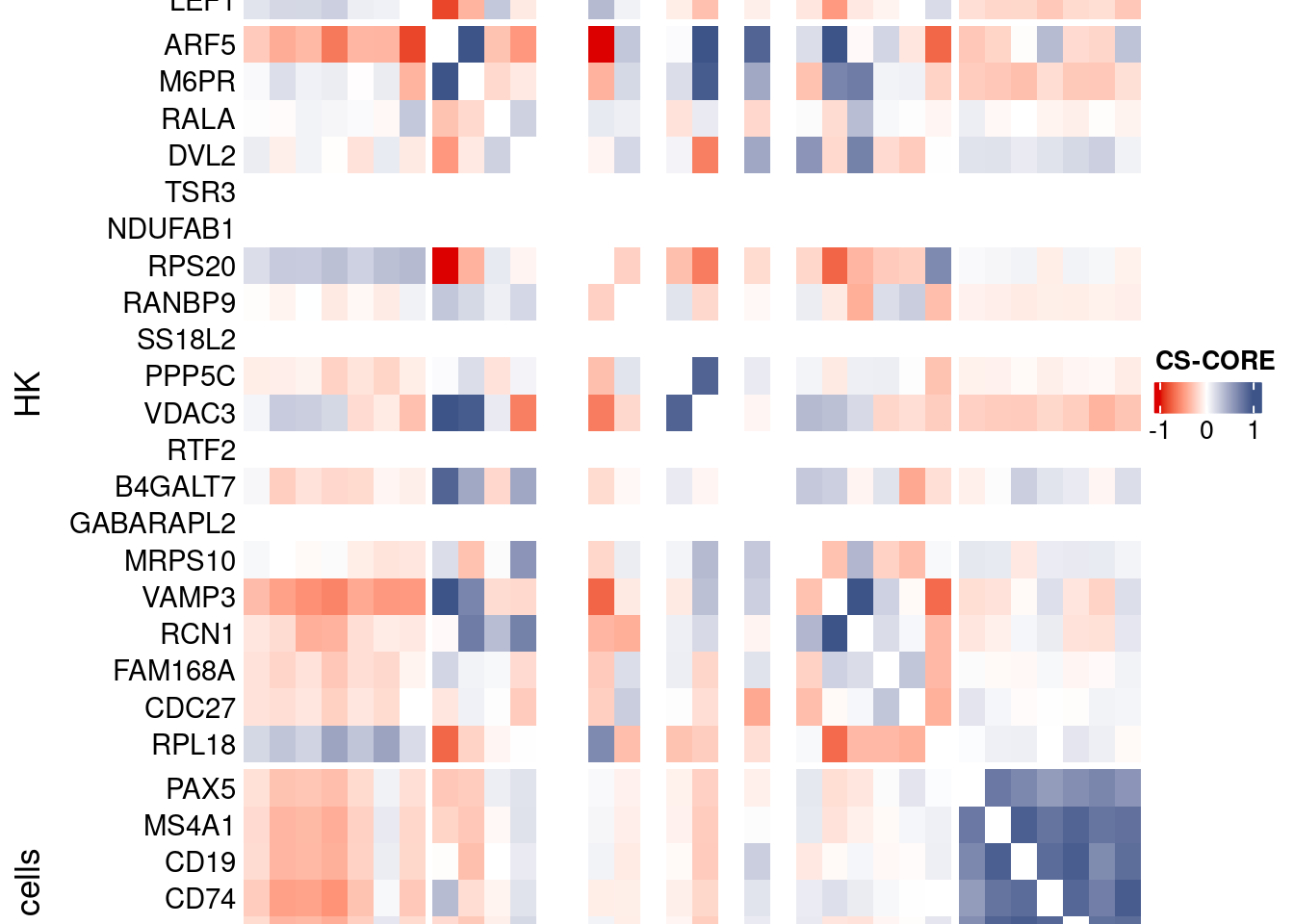
Save CS_CORE matrix
write_fst(as.data.frame(csCoreRes$est), path = paste0("CoexData/CS_CORECorr_", file_code,".fst"),compress = 100)
write_fst(as.data.frame(csCoreRes$p_value), path = paste0("CoexData/CS_COREPValues_", file_code,".fst"),compress = 100)
write.csv(as.data.frame(csCoreRes$p_value), paste0("CoexData/CS_COREPValues_", file_code,".csv"))Baseline: Spearman on UMI counts
corr.pval.list <- correlation_pvaluesSpearman(data = getRawData(obj),
genesFromListExpressed,
n.cells = getNumCells(obj))
data.cor.big <- as.matrix(Matrix::forceSymmetric(corr.pval.list$data.cor, uplo = "U"))
htmp <- correlation_plot(data.cor.big,
genesList, title="UMI baseline S. corr")
p_values.fromSp.C <- corr.pval.list$p_values
data.cor.bigSp.C <- corr.pval.list$data.cor
rm(corr.pval.list)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 11246116 600.7 17840978 952.9 17840978 952.9
Vcells 435302679 3321.1 1054080062 8042.0 1478343794 11278.9draw(htmp, heatmap_legend_side="right")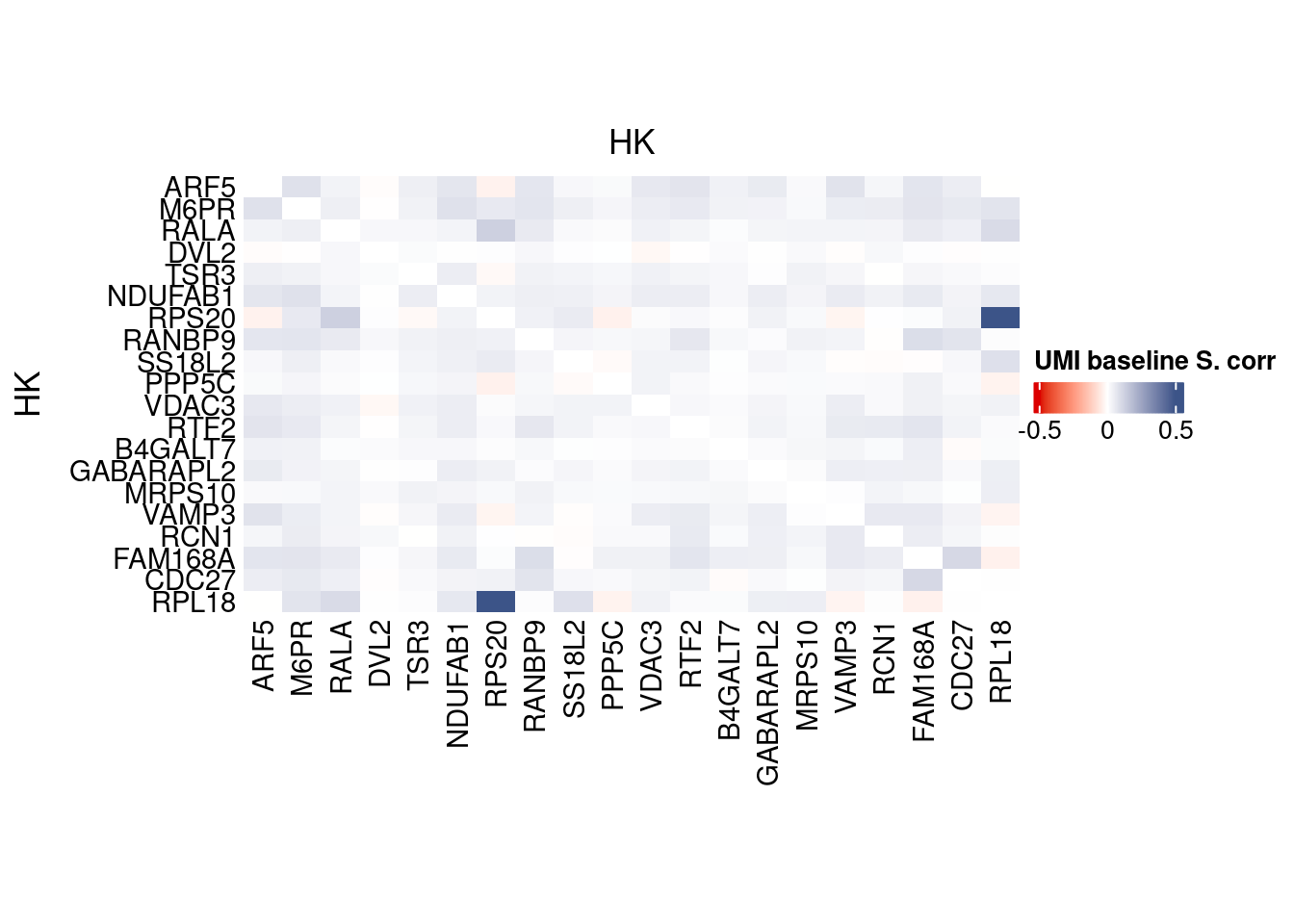
write.csv(as.data.frame(p_values.fromSp.C), paste0("CoexData/BaselineUMISpCorrPValues_", file_code,".csv"))Baseline: Pearson on binarized counts
corr.pval.list <- correlation_pvalues(data = getZeroOneProj(obj),
genesFromListExpressed,
n.cells = getNumCells(obj))
data.cor.big <- as.matrix(Matrix::forceSymmetric(corr.pval.list$data.cor, uplo = "U"))
htmp <- correlation_plotPBMC(data.cor.big,
genesList, title="Zero-one P. corr")
p_values.fromSp.C <- corr.pval.list$p_values
data.cor.bigSp.C <- corr.pval.list$data.cor
rm(corr.pval.list)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 11246174 600.7 17840978 952.9 17840978 952.9
Vcells 435303705 3321.2 1054080062 8042.0 1478343794 11278.9draw(htmp, heatmap_legend_side="right")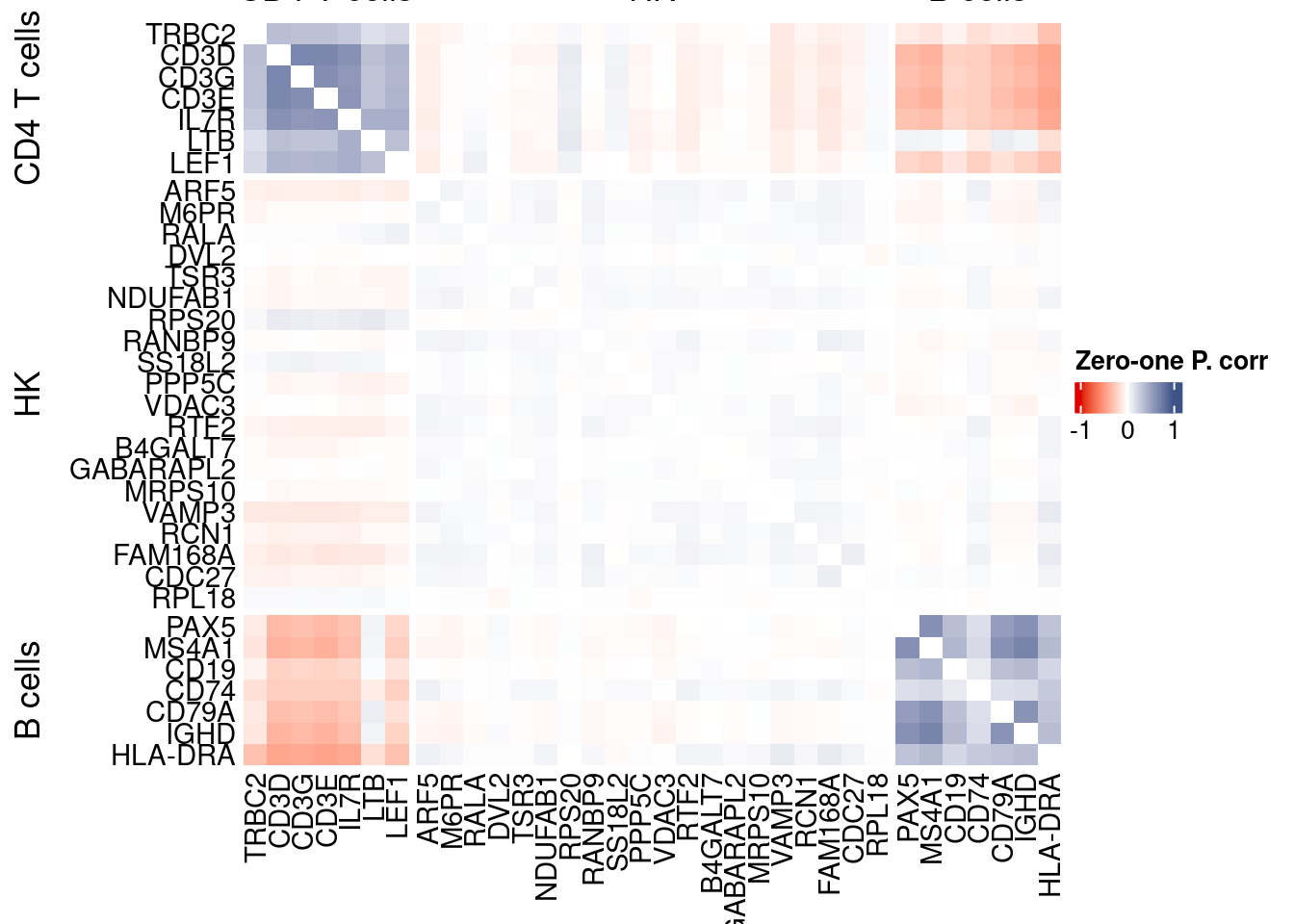
write.csv(as.data.frame(p_values.fromSp.C), paste0("CoexData/ZeroOnePCorrPValues_", file_code,".csv"))Sys.time()[1] "2026-01-22 12:35:55 CET"sessionInfo()R version 4.5.2 (2025-10-31)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 22.04.5 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0 LAPACK version 3.10.0
locale:
[1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
[4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
[7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
time zone: Europe/Rome
tzcode source: system (glibc)
attached base packages:
[1] stats4 parallel grid stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] CSCORE_1.0.2 testthat_3.3.2
[3] monocle3_1.3.7 SingleCellExperiment_1.32.0
[5] SummarizedExperiment_1.38.1 GenomicRanges_1.62.1
[7] Seqinfo_1.0.0 IRanges_2.44.0
[9] S4Vectors_0.48.0 MatrixGenerics_1.22.0
[11] matrixStats_1.5.0 Biobase_2.70.0
[13] BiocGenerics_0.56.0 generics_0.1.3
[15] fstcore_0.10.0 fst_0.9.8
[17] stringr_1.6.0 HiClimR_2.2.1
[19] doParallel_1.0.17 iterators_1.0.14
[21] foreach_1.5.2 Rfast_2.1.5.1
[23] RcppParallel_5.1.10 zigg_0.0.2
[25] Rcpp_1.1.0 patchwork_1.3.2
[27] Seurat_5.4.0 SeuratObject_5.3.0
[29] sp_2.2-0 Hmisc_5.2-3
[31] dplyr_1.1.4 circlize_0.4.16
[33] ComplexHeatmap_2.26.0 COTAN_2.11.1
loaded via a namespace (and not attached):
[1] fs_1.6.6 spatstat.sparse_3.1-0
[3] devtools_2.4.5 httr_1.4.7
[5] RColorBrewer_1.1-3 profvis_0.4.0
[7] tools_4.5.2 sctransform_0.4.2
[9] backports_1.5.0 R6_2.6.1
[11] lazyeval_0.2.2 uwot_0.2.3
[13] ggdist_3.3.3 GetoptLong_1.1.0
[15] urlchecker_1.0.1 withr_3.0.2
[17] gridExtra_2.3 parallelDist_0.2.6
[19] progressr_0.18.0 cli_3.6.5
[21] Cairo_1.7-0 spatstat.explore_3.6-0
[23] fastDummies_1.7.5 labeling_0.4.3
[25] S7_0.2.1 spatstat.data_3.1-9
[27] proxy_0.4-29 ggridges_0.5.6
[29] pbapply_1.7-2 foreign_0.8-90
[31] sessioninfo_1.2.3 parallelly_1.46.0
[33] torch_0.16.3 rstudioapi_0.18.0
[35] shape_1.4.6.1 ica_1.0-3
[37] spatstat.random_3.4-3 distributional_0.6.0
[39] dendextend_1.19.0 Matrix_1.7-4
[41] abind_1.4-8 lifecycle_1.0.4
[43] yaml_2.3.10 SparseArray_1.10.8
[45] Rtsne_0.17 glmGamPoi_1.20.0
[47] promises_1.5.0 crayon_1.5.3
[49] miniUI_0.1.2 lattice_0.22-7
[51] beachmat_2.26.0 cowplot_1.2.0
[53] magick_2.9.0 zeallot_0.2.0
[55] pillar_1.11.1 knitr_1.50
[57] rjson_0.2.23 boot_1.3-32
[59] future.apply_1.20.0 codetools_0.2-20
[61] glue_1.8.0 RcppArmadillo_15.2.3-1
[63] spatstat.univar_3.1-6 remotes_2.5.0
[65] data.table_1.18.0 Rdpack_2.6.4
[67] vctrs_0.7.0 png_0.1-8
[69] spam_2.11-1 gtable_0.3.6
[71] assertthat_0.2.1 cachem_1.1.0
[73] xfun_0.52 rbibutils_2.3
[75] S4Arrays_1.10.1 mime_0.13
[77] reformulas_0.4.1 survival_3.8-3
[79] ncdf4_1.24 ellipsis_0.3.2
[81] fitdistrplus_1.2-2 ROCR_1.0-11
[83] nlme_3.1-168 usethis_3.2.1
[85] bit64_4.6.0-1 RcppAnnoy_0.0.22
[87] rprojroot_2.1.1 GenomeInfoDb_1.44.0
[89] irlba_2.3.5.1 KernSmooth_2.23-26
[91] otel_0.2.0 rpart_4.1.24
[93] colorspace_2.1-1 nnet_7.3-20
[95] tidyselect_1.2.1 processx_3.8.6
[97] bit_4.6.0 compiler_4.5.2
[99] htmlTable_2.4.3 desc_1.4.3
[101] DelayedArray_0.36.0 plotly_4.11.0
[103] checkmate_2.3.2 scales_1.4.0
[105] lmtest_0.9-40 callr_3.7.6
[107] digest_0.6.37 goftest_1.2-3
[109] spatstat.utils_3.2-1 minqa_1.2.8
[111] rmarkdown_2.29 XVector_0.50.0
[113] htmltools_0.5.8.1 pkgconfig_2.0.3
[115] base64enc_0.1-3 coro_1.1.0
[117] lme4_1.1-37 sparseMatrixStats_1.20.0
[119] fastmap_1.2.0 rlang_1.1.7
[121] GlobalOptions_0.1.2 htmlwidgets_1.6.4
[123] ggthemes_5.2.0 UCSC.utils_1.4.0
[125] shiny_1.12.1 DelayedMatrixStats_1.30.0
[127] farver_2.1.2 zoo_1.8-14
[129] jsonlite_2.0.0 BiocParallel_1.44.0
[131] BiocSingular_1.26.1 magrittr_2.0.4
[133] Formula_1.2-5 GenomeInfoDbData_1.2.14
[135] dotCall64_1.2 viridis_0.6.5
[137] reticulate_1.44.1 stringi_1.8.7
[139] brio_1.1.5 MASS_7.3-65
[141] pkgbuild_1.4.7 plyr_1.8.9
[143] listenv_0.10.0 ggrepel_0.9.6
[145] deldir_2.0-4 splines_4.5.2
[147] tensor_1.5 ps_1.9.1
[149] igraph_2.2.1 spatstat.geom_3.6-1
[151] RcppHNSW_0.6.0 pkgload_1.4.0
[153] reshape2_1.4.4 ScaledMatrix_1.16.0
[155] evaluate_1.0.5 nloptr_2.2.1
[157] httpuv_1.6.16 RANN_2.6.2
[159] tidyr_1.3.1 purrr_1.2.0
[161] polyclip_1.10-7 future_1.69.0
[163] clue_0.3-66 scattermore_1.2
[165] ggplot2_4.0.1 rsvd_1.0.5
[167] xtable_1.8-4 RSpectra_0.16-2
[169] later_1.4.2 viridisLite_0.4.2
[171] tibble_3.3.0 memoise_2.0.1
[173] cluster_2.1.8.1 globals_0.18.0